Abstract
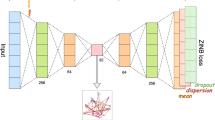
In recent years, with the development of single-cell RNA sequencing (scRNA-seq) technology, more and more scRNA-seq data has been generated. Corresponding analysis methods such as clustering analysis are also proposed, which effectively distinguish the cell types and reveal the cell diversity. However, due to more than ten thousand genes for normal species, the dimension of scRNA-seq data is very high. Meanwhile, there exist many zero counts in scRNA-seq data. They all increase the difficulty of clustering analysis of scRNA-seq data. This paper proposes ScSSC, a semi-supervised clustering method based on 2D embedding. ScSSC uses the autoencoder for pre-training to construct the network and applies the community discovery algorithm to label cells. Then a semi-supervised network is used to clustering the data after training. The clustering results of three public data sets show that ScSSC has better performance than other clustering methods.
N. Shi and Y. Wu—Contributed equally to this work.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Kulkarni, A., Anderson, A.G., Merullo, D.P., Konopka, G.: Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr. Opin. Biotechnol. 58, 129–136 (2019)
Lake, B., Chen, S., Hoshi, M., Plongthongkum, N., Jain, S.: A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 10(1), 2832 (2019)
Lee, E.J., et al.: A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med. 11, 47 (2019). https://doi.org/10.1186/s13073-019-0657-3
Zhang, P., Yang, M., Zhang, Y., Xiao, S., Li, S.: Dissecting the single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 27(6), 1934-1947.e1935 (2019)
Mereu, E., Lafzi, A., Moutinho, C., Ziegenhain, C., Heyn, H.: Benchmarking single-cell RNA sequencing protocols for cell atlas projects. Nature Biotech. 38(6), 747–755 (2020)
Jla, B., Wca, B., Zsa, C.: Single-cell sequencing technologies: current and future. J. Genet. Genomics 41(10), 513–528 (2014)
Xu, C., Su, Z.: Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 12, 1974–1980 (2015)
Wang, B., Zhu, J., Pierson, E., Ramazzotti, D., Batzoglou, S.: Visualization and analysis of single-cell RNA-seq data by kernel-based similarity learning. Nat. Methods 14(4), 414 (2017)
Lin, P., Troup, M., Ho, J.: CIDR: ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol. 18(1), 59 (2017)
Arisdakessian, C., Poirion, O., Yunits, B., Zhu, X., Garmire, L.X.: DeepImpute: an accurate, fast, and scalable deep neural network method to impute single-cell RNA-seq data. Genome Biol. 20(1), 1–14 (2018)
Tian, T., Wan, J., Song, Q., Wei, Z.: Clustering single-cell RNA-seq data with a model-based deep learning approach. Nature Mach. Intell. 1(4), 191 (2019)
Williamson, J.M., Lin, H.M., Lyles, R.H., Hightower, A.W.: Power calculations for ZIP and ZINB models. J. Data Sci. 5(4), 519–534 (2007)
Yue, D., Feng, B., Dai, Q., Wu, L., Altschuler, S.: Massive single-cell RNA-seq analysis and imputation via deep learning (2018)
Li, J., Jiang, W., Han, H., Liu, J., Liu, B., Wang, Y.: ScGSLC: an unsupervised graph similarity learning framework for single-cell RNA-seq data clustering. Comput. Biol. Chem. 90, 107415 (2021)
Schwartz, G.W., Zhou, Y., Petrovic, J., Fasolino, M., Faryabi, R.B.: TooManyCells identifies and visualizes relationships of single-cell clades. Nature Methods 17(4), 1–9 (2020)
Bengio, Y., Courville, A., Vincent, P.: Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1798–1828 (2013)
Ding, J., Condon, A., Shah, S.P.: Interpretable dimensionality reduction of single cell transcriptome data with deep generative models. Nat. Commun. 9(1), 2002 (2018)
Lin, C., Siddhartha, J., Hannah, K., Ziv, B.J.: Using neural networks for reducing the dimensions of single-cell RNA-Seq data. Nucleic Acids Res. 2017(17), e156 (2017)
Guo, X., Long, G., Liu, X., Yin, J.: Improved deep embedded clustering with local structure preservation. In: IJCAI (2017)
Xie, J., Girshick, R., Farhadi, A.: Unsupervised deep embedding for clustering analysis. Computer Science (2015)
Goolam, M., et al.: Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165(1), 61–74 (2016)
Pollen, A., et al.: Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 (2014)
Kolodziejczyk, A.A., et al.: Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17(4), 471–485 (2015)
Nguyen, T.H., Prifti, E., Chevaleyre, Y., Sokolovska, N., Zucker, J.D.: Disease classification in metagenomics with 2D embeddings and deep learning (2018)
Hinton, G.E., Salakhutdinov, R.R.: Reducing the dimensionality of data with neural networks. Science 313(5186), 504–507 (2006)
Blondel, V.D., Guillaume, J.L., Lambiotte, R., Lefebvre, E.: Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 2008(10), P10008 (2008)
Rubinstein, R.: The cross-entropy method for combinatorial and continuous optimization. Methodol. Comput. Appl. Probab. 1(2), 127–190 (1999)
Strehl, A., Ghosh, J.: Cluster ensembles - a knowledge reuse framework for combining multiple partitions. J. Mach. Learn. Res. 3(3), 583–617 (2002)
William, M.: Rand: objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 66(336), 846–850 (1971)
Hubert, L., Arabie, P.: Comparing partitions. J. Classif. 2(1), 193–218 (1985)
Todorov, H., Fournier, D., Gerber, S.: Principal components analysis: theory and application to gene expression data analysis. Genomics Comput. Biol. 4(2), 100041 (2018)
Macqueen, J.: Some methods for classification and analysis of MultiVariate observations. In: Proceedings of Berkeley Symposium on Mathematical Statistics & Probability (1965)
Warren, A.G., Brorson, H., Borud, L.J., Slavin, A.M.D.: A comprehensive review. Ann. Plast. Surg. 59(4), 464–472
Wang, B., Ramazzotti, D., De Sano, L., Zhu, J., Pierson, E.: SIMLR: a tool for large‐scale genomic analyses by multi‐kernel learning. Proteomics 11(3), 333 (2018)
Kiselev, V.Y., et al.: SC3: consensus clustering of single-cell RNA-seq data. Nature Methods 14(5), 483–486 (2017)
Baran, P.M., Dennis, K.: Random forest based similarity learning for single cell RNA sequencing data. Bioinformatics 13, i79–i88 (2018)
Pyramid Match Kernels: Discriminative Classification with Sets of Image Features (2006)
Grauman, K., Darrell, T.: The pyramid match kernel: discriminative classification with sets of image features. In: Tenth IEEE International Conference on Computer Vision (2005)
Laurens, V.D.M., Hinton, G.: Visualizing data using t-SNE. J. Mach. Learn. Res. 9(2605), 2579–2605 (2008)
Acknowledgements
This work was supported by the grants from the National Key Research Program (2017YFC1201201, 2018YFC0910504 and 2017YFC0907503), Shenzhen Science and Technology Program the university stable support program (20200821222112001) from JL.
Funding
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Contributions
NS, YW and JL designed the study, performed bioinformatics analysis and drafted the manuscript. All of the authors performed the analysis and participated in the revision of the manuscript. JL conceived of the study, participated in its design and coordination and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
All additional files are available at: https://github.com/NaiLeShi/-ScSSC.
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Shi, N., Wu, Y., Du, L., Liu, B., Wang, Y., Li, J. (2021). ScSSC: Semi-supervised Single Cell Clustering Based on 2D Embedding. In: Huang, DS., Jo, KH., Li, J., Gribova, V., Premaratne, P. (eds) Intelligent Computing Theories and Application. ICIC 2021. Lecture Notes in Computer Science(), vol 12838. Springer, Cham. https://doi.org/10.1007/978-3-030-84532-2_43
Download citation
DOI: https://doi.org/10.1007/978-3-030-84532-2_43
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-84531-5
Online ISBN: 978-3-030-84532-2
eBook Packages: Computer ScienceComputer Science (R0)