Abstract
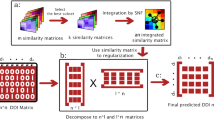
The experimental determination of drug-target interaction is time-consuming and expensive. Therefore, a continuous demand for more effective prediction of drug-target interaction using computing technology. Many algorithms have been designed to infer potential interactions. Most of these algorithms rely on drug similarity and target similarity as auxiliary information in modeling, but they ignore the problem that there are a lot of missing auxiliary data of existing drugs or targets, which affects the prediction performance of the model and fails to achieve the expected effect. Here, we propose a calculation model named MCLO, which is based on the matrix completion and linear optimization technology to predict novel drug-target interactions. First, the proposed method calculate the side effect similarity of drugs and protein-protein interaction similarity of targets. Then we utilize the idea of linear neighbor representation learning to predict the potential drug-target interaction. It is worth mentioning that our method uses the idea of matrix completion technology to complete the imperfect similarity before drug-target interactions prediction. To evaluate the performance of MCLO, we carry out experiments on four gold standard datasets. The experimental results show that MCLO can be effectively applied to identify drug-target interactions.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Olayan, R.S., Ashoor, H., Bajic, V.B.: DDR: efficient computational method to predict drug–target interactions using graph mining and machine learning approaches. Bioinformatics 34(7), 1164–1173 (2017)
Goh, K.I., Cusick, M.E., Valle, D., et al.: The human disease network. Proc. Natl. Acad. Sci. 104(21), 8685–8690 (2007)
Avorn, J.: The $2.6 billion pill--methodologic and policy considerations. New Engl. J. Med. 372(20), 1877–1879 (2015)
Ming, H., Bryant, S.H., Wang, Y.: Predicting drug-target interactions by dual-network integrated logistic matrix factorization. Sci. Rep. 7, 40376 (2017)
Malina, D., Greene, J.A., Loscalzo, J.: Putting the patient back together - social medicine, network medicine, and the limits of reductionism. New Engl. J. Med. 377(25), 2493 (2017)
Guo, L., Yan, Z., Zheng, X., et al.: A comparison of various optimization algorithms of protein–ligand docking programs by fitness accuracy. J. Mol. Model. 20(7), 2251 (2014). https://doi.org/10.1007/s00894-014-2251-3
Liu, Y., et al.: Neighborhood regularized logistic matrix factorization for drug-target interaction prediction. PLoS Comput. Biol. 12 (2016)
Peng, J., Li, J., Shang, X.: A learning-based method for drug-target interaction prediction based on feature representation learning and deep neural network. BMC Bioinform. 21(Suppl 13), 394 (2020)
Yamanishi, Y., Kotera, M., Kanehisa, M., Goto, S.: Drug-target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics 26(12), 246–254 (2010)
Huang, Y., et al.: Predicting drug-target on heterogeneous network with co-rank. In: The 8th International Conference on Computer Engineering and Networks, pp. 571–81 (2020)
Cheng, F., et al.: Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol. 8(5), 1002503 (2012)
Zzat, A.E., Zhao, P., Min, W., et al.: Drug-target interaction prediction with graph regularized matrix factorization. IEEE/ACM Trans. Comput. Biol. Bioinform. 14(3), 1 (2016)
Yamanishi, Y., Araki, M., Gutteridge, A., Honda, W., Kanehisa, M.: Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 24(13), i232–i240 (2008)
Mei, J.-P., Kwoh, C.-K., Yang, P., Li, X.-L., Zheng, J.: Drug–target interaction prediction by learning from local information and neighbors, Bioinformatics 29(2), 238–245 (2013)
Ba-Alawi, W., et al.: DASPfind: new efficient method to predict drug-target interactions. Cheminform 8(1), 15 (2016)
Daminelli, S., et al.: Common neighbours and the local-community-paradigm for topological link prediction in bipartite networks. New J. Phys. 17(11), 113037 (2015)
Kanehisa, M., et al.: From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34, D354–D357 (2006)
Gunther, S., et al.: Super target and matador: resources for exploring drug-target relationships. Nucleic Acids Res. 36, D919–D922 (2008)
Wishart, D.S., et al.: DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–D906 (2008)
Schomburg, I.: BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 32, D431–D433 (2004)
Kuhn, M., Letunic, I., Jensen, L.J., et al.: The SIDER database of drugs and side effects. Nucleic Acids Res. 44(D1), D1075–D1079 (2016)
Alanis-Lobato, G., Andrade-Navarro, M.A., Schaefer, M.H.: HIPPIE v2.0: enhancing meaningfulness and reliability of protein–protein interaction networks. Nucleic Acids Res. 45(D1), D408–D414 (2017)
Campillos, M., Kuhn, M., Gavin, A.C., et al.: Drug target identification using side-effect similarity. Science 321(5886), 263–266 (2008)
Vilar, S., Hripcsak, G.: The role of drug profiles as similarity metrics: applications to repurposing, adverse effects detection and drug-drug interactions. Brief. Bioinform. 18(4), bbw048 (2016)
Deng, M., et al.: Prediction of protein function using protein-protein interaction data. In: Proceedings IEEE Computer Society Bioinformatics Conference EE (2002)
Shen, C., Luo, J., Lai, Z., et al.: Multiview joint learning-based method for identifying small-molecule-associated MiRNAs by integrating pharmacological, genomics, and network knowledge. J. Chem. Inf. Model. (2020)
Lin, Z., Chen, M., Ma, Y.: The augmented Lagrange multiplier method for exact recovery of corrupted low-rank matrices. eprint arxiv:9 (2010)
Pech, R., Hao, D., Lee, Y.L., et al.: Link prediction via linear optimization. Phys. A: Stat. Mech. Appl. 528 (2019)
van Laarhoven, T., Marchiori, E.: Predicting drug-target interactions for new drug compounds using a weighted nearest neighbor profile. PLoS ONE 8(6), e66952 (2013)
Zheng, X., Ding, H., Mamitsuka, H., Zhu, S.: Collaborative matrix factorization with multiple similarities for predicting drug-target interactions. In: KDD 2013: Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1025–1033 (2013)
Liu, Y., et al.: Neighborhood regularized logistic matrix factorization for drug-target interaction prediction. PLoS Comput. Biol. 12 e1004760 (2016)
Ding, Y., Tang, J., Guo, F.: Identification of drug-target interactions via dual Laplacian regularized least squares with multiple Kernel fusion. Knowl.-Based Syst. 204, 106254 (2020)
Acknowledgements
This work was supported by Natural Science Foundation of China (Grant No. 61972141) and Natural Science Foundation of Hunan Province, China (Grant No. 2020JJ4209).
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Lu, X., Liu, F., Li, J., He, K., Jiang, K., Gu, C. (2021). An Efficient Computational Method to Predict Drug-Target Interactions Utilizing Matrix Completion and Linear Optimization Method. In: Huang, DS., Jo, KH., Li, J., Gribova, V., Premaratne, P. (eds) Intelligent Computing Theories and Application. ICIC 2021. Lecture Notes in Computer Science(), vol 12838. Springer, Cham. https://doi.org/10.1007/978-3-030-84532-2_54
Download citation
DOI: https://doi.org/10.1007/978-3-030-84532-2_54
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-84531-5
Online ISBN: 978-3-030-84532-2
eBook Packages: Computer ScienceComputer Science (R0)