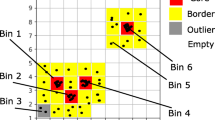
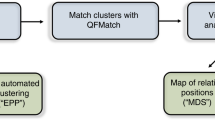
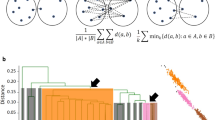
Abstract
Hierarchical clustering is a common tool for simplification, exploration, and analysis of datasets in many areas of research. For data originating in flow cytometry, a specific variant of agglomerative clustering based Mahalanobis-average linkage has been shown to produce results better than the common linkages. However, the high complexity of computing the distance limits the applicability of the algorithm to datasets obtained from current equipment. We propose an optimized, GPU-accelerated open-source implementation of the Mahalanobis-average hierarchical clustering that improves the algorithm performance by over two orders of magnitude, thus allowing it to scale to the large datasets. We provide a detailed analysis of the optimizations and collected experimental results that are also portable to other hierarchical clustering algorithms; and demonstrate the use on realistic high-dimensional datasets.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
CUDA C++ Programming Guide (2021). https://docs.nvidia.com/cuda/cuda-c-programming-guide
Bodenheimer, T., et al.: Fastpg: fast clustering of millions of single cells. bioRxiv (2020)
Chang, D.J., Kantardzic, M.M., Ouyang, M.: Hierarchical clustering with cuda/gpu. In: ISCA PDCCS, pp. 7–12. Citeseer (2009)
Cuomo, S., De Angelis, V., Farina, G., Marcellino, L., Toraldo, G.: A gpu-accelerated parallel k-means algorithm. Comput. Electric. Eng. 75, 262–274 (2019)
Everitt, B., Skrondal, A.: The Cambridge dictionary of statistics, vol. 106. Cambridge University Press, Cambridge (2002)
Fišer, K., et al.: Detection and monitoring of normal and leukemic cell populations with hierarchical clustering of flow cytometry data. Cytometry Part A 81(1), 25–34 (2012)
van Gassen, S., et al.: Flowsom: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry Part A 87(7), 636–645 (2015)
Gowanlock, M., Rude, C.M., Blair, D.M., Li, J.D., Pankratius, V.: Clustering throughput optimization on the gpu. In: 2017 IEEE International Parallel and Distributed Processing Symposium (IPDPS), pp. 832–841. IEEE (2017)
Kratochvíl, M., Bednárek, D., Sieger, T., Fišer, K., Vondrášek, J.: Shinysom: graphical som-based analysis of single-cell cytometry data. Bioinformatics 36(10), 3288–3289 (2020)
Kruliš, M., Kratochvíl, M.: Detailed analysis and optimization of cuda k-means algorithm. In: 49th International Conference on Parallel Processing-ICPP, pp. 1–11 (2020)
Lugli, E., Roederer, M., Cossarizza, A.: Data analysis in flow cytometry: the future just started. Cytometry Part A 77(7), 705–713 (2010)
Mahalanobis, P.C.: On the generalized distance in statistics. National Institute of Science of India (1936)
Shapiro, H.M.: Practical Flow Cytometry. John Wiley & Sons, Hoboken (2005)
Shirkhorshidi, A.S., Aghabozorgi, S., Wah, T.Y.: A comparison study on similarity and dissimilarity measures in clustering continuous data. PloS one 10(12), e0144059 (2015)
Weber, L.M., Robinson, M.D.: Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry Part A 89(12), 1084–1096 (2016)
Zhang, Q., Zhang, Y.: Hierarchical clustering of gene expression profiles with graphics hardware acceleration. Pattern Recogn. Lett. 27(6), 676–681 (2006)
Acknowledgements
This work was supported by Czech Science Foundation (GAČR) project 19-22071Y, by ELIXIR CZ LM2018131 (MEYS), by Charles University grant SVV-260451, and by Czech Health Research Council (AZV) [NV18-08-00385].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Šmelko, A., Kratochvíl, M., Kruliš, M., Sieger, T. (2021). GPU-Accelerated Mahalanobis-Average Hierarchical Clustering Analysis. In: Sousa, L., Roma, N., Tomás, P. (eds) Euro-Par 2021: Parallel Processing. Euro-Par 2021. Lecture Notes in Computer Science(), vol 12820. Springer, Cham. https://doi.org/10.1007/978-3-030-85665-6_36
Download citation
DOI: https://doi.org/10.1007/978-3-030-85665-6_36
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-85664-9
Online ISBN: 978-3-030-85665-6
eBook Packages: Computer ScienceComputer Science (R0)