Abstract
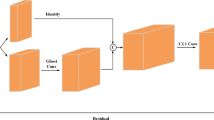
Datasets from real-world applications usually deal with many variables and present difficulties when modeling them with traditional classifiers. There is a variety of feature selection and extraction tools that may help with the dimensionality problem, but most of them do not focus on the complexity of the classes. In this paper, a new autoencoder-based model for addressing class complexity in data is introduced, aiming to extract features that present classes in a more separable fashion, thus simplifying the classification task. This is possible thanks to a combination of the standard reconstruction error with a least-squares support vector machine loss function. This model is then applied to a practical use case: classification of chest X-rays according to the presence of COVID-19, showing that learning features that increase linear class separability can boost classification performance. For this purpose, a specific convolutional autoencoder architecture has been designed and trained using the recently published COVIDGR dataset. The proposed model is evaluated by means of several traditional classifiers and metrics, in order to establish the improvements caused by the extracted features. The advantages of using a feature learner and traditional classifiers are also discussed.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abadi, M., et al.: TensorFlow: large-scale machine learning on heterogeneous systems (2015). https://www.tensorflow.org/
Afshar, P., Heidarian, S., Naderkhani, F., Oikonomou, A., Plataniotis, K.N., Mohammadi, A.: COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from x-ray images. Pattern Recogn. Lett. 138, 638–643 (2020)
Aggarwal, C.C.: Data Classification, pp. 285–344. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-14142-8_10
Aggarwal, C.C., Hinneburg, A., Keim, D.A.: On the surprising behavior of distance metrics in high dimensional space. In: Van den Bussche, J., Vianu, V. (eds.) ICDT 2001. LNCS, vol. 1973, pp. 420–434. Springer, Heidelberg (2001). https://doi.org/10.1007/3-540-44503-X_27
Basu, M., Ho, T.K.: Data Complexity in Pattern Recognition. Springer, Heidelberg (2006). https://doi.org/10.1007/978-1-84628-172-3
Beyer, K., Goldstein, J., Ramakrishnan, R., Shaft, U.: When is “nearest neighbor’’ meaningful? In: Beeri, C., Buneman, P. (eds.) ICDT 1999. LNCS, vol. 1540, pp. 217–235. Springer, Heidelberg (1999). https://doi.org/10.1007/3-540-49257-7_15
Charte, D., Charte, F., del Jesus, M.J., Herrera, F.: An analysis on the use of autoencoders for representation learning: fundamentals, learning task case studies, explainability and challenges. Neurocomputing 404, 93–107 (2020). https://doi.org/10.1016/j.neucom.2020.04.057
Charte, D., Charte, F., García, S., del Jesus, M.J., Herrera, F.: A practical tutorial on autoencoders for nonlinear feature fusion: taxonomy, models, software and guidelines. Inform. Fusion 44, 78–96 (2018). https://doi.org/10.1016/j.inffus.2017.12.007
García, S., Luengo, J., Herrera, F.: Data Preprocessing in Data Mining, vol. 72. Springer, Heidelberg (2015). https://doi.org/10.1007/978-3-319-10247-4
Gong, J., et al.: A tool for early prediction of severe coronavirus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis. 71(15), 833–840 (2020)
He, K., Zhang, X., Ren, S., Sun, J.: Identity mappings in deep residual networks. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9908, pp. 630–645. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46493-0_38
Knight, S.R., et al.: Risk stratification of patients admitted to hospital with COVID-19 using the ISARIC WHO clinical characterisation protocol: development and validation of the 4C mortality score. bmj 370, 1–13 (2020)
Liu, X., et al.: Self-supervised learning: generative or contrastive. arXiv preprint arXiv:2006.082181(2) (2020)
Luengo, J., Fernández, A., García, S., Herrera, F.: Addressing data complexity for imbalanced data sets: analysis of smote-based oversampling and evolutionary undersampling. Soft. Comput. 15(10), 1909–1936 (2011)
Maguolo, G., Nanni, L.: A critic evaluation of methods for COVID-19 automatic detection from x-ray images. Inform. Fusion 76, 1–7 (2021). https://doi.org/10.1016/j.inffus.2021.04.008
Makhzani, A., Shlens, J., Jaitly, N., Goodfellow, I., Frey, B.: Adversarial autoencoders. arXiv preprint arXiv:1511.05644 (2015)
Pascual-Triana, J.D., Charte, D., Arroyo, M.A., Fernández, A., Herrera, F.: Revisiting data complexity metrics based on morphology for overlap and imbalance: snapshot, new overlap number of balls metrics and singular problems prospect. Knowl. Inf. Syst. 63, 1961–1989 (2021)
Suykens, J.A., Vandewalle, J.: Least squares support vector machine classifiers. Neural Process. Lett. 9(3), 293–300 (1999)
Tabik, S., Gómez-Ríos, A., Martín-Rodríguez, J.L., Sevillano-García, I., Rey-Area, M., Charte, D., et al.: COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest x-ray images. IEEE J. Biomed. Health Inform. 24(12), 3595–3605 (2020). https://doi.org/10.1109/JBHI.2020.3037127
Wang, L.: Feature selection with kernel class separability. IEEE Trans. Pattern Anal. Mach. Intell. 30(9), 1534–1546 (2008)
Wang, L., Lin, Z.Q., Wong, A.: COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci. Rep. 10(1), 1–12 (2020)
Yu, X., Chen, Y., Li, T., Liu, S., Li, G.: Multi-mapping image-to-image translation via learning disentanglement. arXiv preprint arXiv:1909.07877 (2019)
Zhang, Y., Li, S., Wang, T., Zhang, Z.: Divergence-based feature selection for separate classes. Neurocomputing 101, 32–42 (2013). https://doi.org/10.1016/j.neucom.2012.06.036
Acknowledgments
D. Charte is supported by the Spanish Ministry of Science under the FPU National Program (Ref. FPU17/04069). F. Charte is supported by the Spanish Ministry of Science project PID2019-107793GB-I00/AEI/10.13039/501100011033. F. Herrera is supported by the Spanish Ministry of Science project PID2020-119478GB-I00 and the Andalusian Excellence project P18-FR-4961. This work is supported by the project COVID19RX-Ayudas Fundación BBVA a Equipos de Investigación Científica SARS-CoV-2 y COVID-19 2020.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Charte, D., Sevillano-García, I., Lucena-González, M.J., Martín-Rodríguez, J.L., Charte, F., Herrera, F. (2021). Slicer: Feature Learning for Class Separability with Least-Squares Support Vector Machine Loss and COVID-19 Chest X-Ray Case Study. In: Sanjurjo González, H., Pastor López, I., García Bringas, P., Quintián, H., Corchado, E. (eds) Hybrid Artificial Intelligent Systems. HAIS 2021. Lecture Notes in Computer Science(), vol 12886. Springer, Cham. https://doi.org/10.1007/978-3-030-86271-8_26
Download citation
DOI: https://doi.org/10.1007/978-3-030-86271-8_26
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-86270-1
Online ISBN: 978-3-030-86271-8
eBook Packages: Computer ScienceComputer Science (R0)