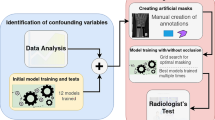
Abstract
Convolutional neural networks are showing promise in the automatic diagnosis of thoracic pathologies on chest x-rays. Their black-box nature has sparked many recent works to explain the prediction via input feature attribution methods (aka saliency methods). However, input feature attribution methods merely identify the importance of input regions for the prediction and lack semantic interpretation of model behavior. In this work, we first identify the semantics associated with internal units (feature maps) of the network. We proceed to investigate the following questions; Does a regression model that is only trained with COVID-19 severity scores implicitly learn visual patterns associated with thoracic pathologies? Does a network that is trained on weakly labeled data (e.g. healthy, unhealthy) implicitly learn pathologies? Moreover, we investigate the effect of pretraining and data imbalance on the interpretability of learned features. In addition to the analysis, we propose semantic attribution to semantically explain each prediction. We present our findings using publicly available chest pathologies (CheXpert [5], NIH ChestX-ray8 [25]) and COVID-19 datasets (BrixIA [20], and COVID-19 chest X-ray segmentation dataset [4]). The Code (https://github.com/CAMP-eXplain-AI/CheXplain-Dissection) is publicly available.
S. T. Kim and N. Navab shared senior authorship.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bach, S., Binder, A., Montavon, G., Klauschen, F., Müller, K.R., Samek, W.: On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 10(7), e0130140 (2015)
Bau, D., Zhou, B., Khosla, A., Oliva, A., Torralba, A.: Network dissection: quantifying interpretability of deep visual representations. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6541–6549 (2017)
Huang, G., Liu, Z., Weinberger, K.Q.: Densely connected convolutional networks. CoRR abs/1608.06993 (2016). http://arxiv.org/abs/1608.06993
Inc, G.B.: Covid-19 chest x-ray segmentations dataset. https://github.com/GeneralBlockchain/covid-19-chest-xray-segmentations-dataset
Irvin, J., et al.: Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 590–597 (2019)
Johnson, A.E., et al.: MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv preprint arXiv:1901.07042 (2019)
Karim, M., Döhmen, T., Rebholz-Schuhmann, D., Decker, S., Cochez, M., Beyan, O., et al.: Deepcovidexplainer: Explainable covid-19 predictions based on chest x-ray images. arXiv preprint arXiv:2004.04582 (2020)
Khakzar, A., Albarqouni, S., Navab, N.: Learning interpretable features via adversarially robust optimization. In: Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 793–800. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_88
Khakzar, A., Baselizadeh, S., Khanduja, S., Rupprecht, C., Kim, S.T., Navab, N.: Neural response interpretation through the lens of critical pathways. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2021)
Khakzar, A., et al.: Explaining COVID-19 and thoracic pathology model predictions by identifying informative input features (2021)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Li, Z., et al.: Thoracic disease identification and localization with limited supervision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8290–8299 (2018)
Nguyen, A., Dosovitskiy, A., Yosinski, J., Brox, T., Clune, J.: Synthesizing the preferred inputs for neurons in neural networks via deep generator networks. In: Advances in Neural Information Processing Systems, pp. 3387–3395 (2016)
Oh, Y., Park, S., Ye, J.C.: Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging 39(8), 2688–2700 (2020)
Olah, C., Mordvintsev, A., Schubert, L.: Feature visualization. Distill (2017). https://doi.org/10.23915/distill.00007, https://distill.pub/2017/feature-visualization
Punn, N.S., Agarwal, S.: Automated diagnosis of COVID-19 with limited posteroanterior chest X-ray images using fine-tuned deep neural networks. Appl. Intell. 51(5), 2689–2702 (2020). https://doi.org/10.1007/s10489-020-01900-3
Rajpurkar, P., et al.: Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225 (2017)
Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D.: Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017)
Shapley, L.S.: A value for n-person games. Contrib. Theor. Games 2(28), 307–317 (1953)
Signoroni, A., et al.: End-to-end learning for semiquantitative rating of COVID-19 severity on chest X-rays. arXiv preprint arXiv:2006.04603 (2020)
Simonyan, K., Vedaldi, A., Zisserman, A.: Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034 (2013)
Sundararajan, M., Najmi, A.: The many Shapley values for model explanation. In: 37th International Conference on Machine Learning, ICML 2020 (2020)
Sundararajan, M., Taly, A., Yan, Q.: Axiomatic attribution for deep networks. In: International Conference on Machine Learning, pp. 3319–3328. PMLR (2017)
Taghanaki, S.A., et al.: InfoMask: Masked Variational Latent Representation to Localize Chest Disease. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 739–747. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_82
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., Summers, R.M.: Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2097–2106 (2017)
Wu, J., et al.: Deepminer: discovering interpretable representations for mammogram classification and explanation. arXiv preprint arXiv:1805.12323 (2018)
Zeiler, M.D., Fergus, R.: Visualizing and understanding convolutional networks. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8689, pp. 818–833. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10590-1_53
Acknowledgement
This work is partially funded by the Munich Center for Machine Learning (MCML) and the Bavarian Research Foundation grant AZ-1429-20C. The computational resources for the study are provided by the Amazon Web Services Diagnostic Development Initiative. S.T. Kim is supported by the Korean MSIT, under the National Program for Excellence in SW (2017-0-00093), supervised by the IITP.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Khakzar, A. et al. (2021). Towards Semantic Interpretation of Thoracic Disease and COVID-19 Diagnosis Models. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12903. Springer, Cham. https://doi.org/10.1007/978-3-030-87199-4_47
Download citation
DOI: https://doi.org/10.1007/978-3-030-87199-4_47
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87198-7
Online ISBN: 978-3-030-87199-4
eBook Packages: Computer ScienceComputer Science (R0)