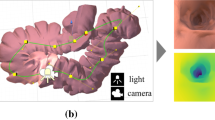
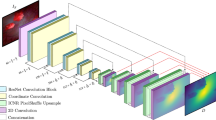
Abstract
Depth estimation in colonoscopy images provides geometric clues for downstream medical analysis tasks, such as polyp detection, 3D reconstruction, and diagnosis. Recently, deep learning technology has made significant progress in monocular depth estimation for natural scenes. However, without sufficient ground truth of dense depth maps for colonoscopy images, it is significantly challenging to train deep neural networks for colonoscopy depth estimation. In this paper, we propose a novel approach that makes full use of both synthetic data and real colonoscopy videos. We use synthetic data with ground truth depth maps to train a depth estimation network with a generative adversarial network model. Despite the lack of ground truth depth, real colonoscopy videos are used to train the network in a self-supervision manner by exploiting temporal consistency between neighboring frames. Furthermore, we design a masked gradient warping loss in order to ensure temporal consistency with more reliable correspondences. We conducted both quantitative and qualitative analysis on an existing synthetic dataset and a set of real colonoscopy videos, demonstrating the superiority of our method on more accurate and consistent depth estimation for colonoscopy images.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Arnold, M., Sierra, M.S., Laversanne, M., Soerjomataram, I., Jemal, A., Bray, F.: Global patterns and trends in colorectal cancer incidence and mortality. Gut 66(4), 683–691 (2017). https://doi.org/10.1136/gutjnl-2015-310912
Freedman, D., Blau, Y., Katzir, L., Aides, A., Shimshoni, I., Veikherman, D., Golany, T., Gordon, A., Corrado, G., Matias, Y., Rivlin, E.: Detecting deficient coverage in colonoscopies. IEEE Trans. Med. Imag. 39(11), 3451–3462 (2020). https://doi.org/10.1109/TMI.2020.2994221
Hong, D., Tavanapong, W., Wong, J., Oh, J., de Groen, P.C.: 3D reconstruction of virtual colon structures from colonoscopy images. Comput. Med. Imag. Graph. 38(1), 22–33 (2014). https://doi.org/10.1016/j.compmedimag.2013.10.005
Isola, P., Zhu, J., Zhou, T., Efros, A.A.: Image-to-image translation with conditional adversarial networks. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 5967–5976 (2017). https://doi.org/10.1109/CVPR.2017.632
Itoh, H., et al.: Towards automated colonoscopy diagnosis: Binary polyp size estimation via unsupervised depth learning. In: Medical Image Computing and Computer Assisted (MICCAI 2018), pp. 611–619 (2018)
Liu, X., Sinha, A., Ishii, M., Hager, G.D., Reiter, A., Taylor, R.H., Unberath, M.: Dense depth estimation in monocular endoscopy with self-supervised learning methods. IEEE Trans. Med. Imag. 39(5), 1438–1447 (2020). https://doi.org/10.1109/TMI.2019.2950936
Ma, R., Wang, R., Pizer, S., Rosenman, J., McGill, S.K., Frahm, J.M.: Real-time 3D reconstruction of colonoscopic surfaces for determining missing regions. In: Medical Image Computing and Computer Assisted Intervention, pp. 573–582 (2019). https://doi.org/10.1007/978-3-030-32254-0_64
Mahmood, F., Durr, N.J.: Deep learning and conditional random fields-based depth estimation and topographical reconstruction from conventional endoscopy. Med. Image Anal. 48, 230–243 (2018). https://doi.org/10.1016/j.media.2018.06.005
Nadeem, S., Kaufman, A.: Depth reconstruction and computer-aided polyp detection in optical colonoscopy video frames. arXiv preprint arXiv:1609.01329 (2016)
Odena, A., Dumoulin, V., Olah, C.: Deconvolution and checkerboard artifacts. Distill (2016). https://doi.org/10.23915/distill.00003
Rau, A., Edwards, P.E., Ahmad, O.F., Riordan, P., Janatka, M., Lovat, L.B., Stoyanov, D.: Implicit domain adaptation with conditional generative adversarial networks for depth prediction in endoscopy. Int. J. Comput. Assist. Radiol. Surg. 14(7), 1167–1176 (2019). https://doi.org/10.1007/s11548-019-01962-w
Schonberger, J.L., Frahm, J.: Structure-from-motion revisited. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 4104–4113 (2016). https://doi.org/10.1109/CVPR.2016.445
Stark, U.A., Frese, T., Unverzagt, S., Bauer, A.: What is the effectiveness of various invitation methods to a colonoscopy in the early detection and prevention of colorectal cancer? protocol of a systematic review. Syst. Rev. 9(1), 1–7 (2020). https://doi.org/10.1186/s13643-020-01312-x
Sun, D., Yang, X., Liu, M., Kautz, J.: PWC-net: CNNs for optical flow using pyramid, warping, and cost volume. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 8934–8943 (2018). https://doi.org/10.1109/CVPR.2018.00931
Waluga, M., Zorniak, M., Fichna, J., Kukla, M., Hartleb, M.: Pharmacological and dietary factors in prevention of colorectal cancer. J. Physiol. Pharmacol. 69(3) (2018). https://doi.org/10.26402/jpp.2018.3.02
Wang, T., Liu, M., Zhu, J., Tao, A., Kautz, J., Catanzaro, B.: High-resolution image synthesis and semantic manipulation with conditional GANs. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 8798–8807 (2018). https://doi.org/10.1109/CVPR.2018.00917
Widya, A.R., Monno, Y., Okutomi, M., Suzuki, S., Gotoda, T., Miki, K.: Stomach 3D reconstruction based on virtual chromoendoscopic image generation. In: The 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC), pp. 1848–1852 (2020). https://doi.org/10.1109/EMBC44109.2020.9176016
Widya, A.R., Monno, Y., Okutomi, M., Suzuki, S., Gotoda, T., Miki, K.: Whole stomach 3D reconstruction and frame localization from monocular endoscope video. IEEE J. Trans. Eng. Health Med. 7, 1–10 (2019). https://doi.org/10.1109/JTEHM.2019.2946802
Widya, A.R., Monno, Y., Okutomi, M., Suzuki, S., Gotoda, T., Miki, K.: Self-supervised monocular depth estimation in gastroendoscopy using GAN-augmented images. In: Medical Imaging 2021: Image Processing. vol. 11596, p. 1159616 (2021). https://doi.org/10.1117/12.2579317
Yu, L., Chen, H., Dou, Q., Qin, J., Heng, P.A.: Integrating online and offline three-dimensional deep learning for automated polyp detection in colonoscopy videos. IEEE J. Biomed. Health Inform. 21(1), 65–75 (2017). https://doi.org/10.1109/JBHI.2016.2637004
Zhang, R., Zheng, Y., Poon, C.C., Shen, D., Lau, J.Y.: Polyp detection during colonoscopy using a regression-based convolutional neural network with a tracker. Patt. Recogn. 83, 209–219 (2018). https://doi.org/10.1016/j.patcog.2018.05.026
Zhao, Q., Price, T., Pizer, S., Niethammer, M., Alterovitz, R., Rosenman, J.: The endoscopogram: A 3D model reconstructed from endoscopic video frames. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI 2016), pp. 439–447 (2016). https://doi.org/10.1007/978-3-319-46720-7_51
Zhou, T., Brown, M., Snavely, N., Lowe, D.G.: Unsupervised learning of depth and ego-motion from video. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 6612–6619 (2017). https://doi.org/10.1109/CVPR.2017.700
Acknowledgments
We acknowledge funding from National Natural Science Foundation of China under Grants 61976007 and 62076230.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (mp4 36725 KB)
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Cheng, K., Ma, Y., Sun, B., Li, Y., Chen, X. (2021). Depth Estimation for Colonoscopy Images with Self-supervised Learning from Videos. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12906. Springer, Cham. https://doi.org/10.1007/978-3-030-87231-1_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-87231-1_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87230-4
Online ISBN: 978-3-030-87231-1
eBook Packages: Computer ScienceComputer Science (R0)