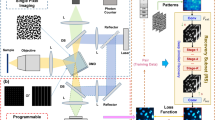
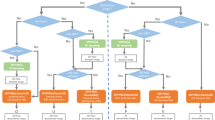
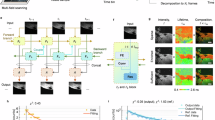
Abstract
Compressed sensing fluorescence microscopy (CS-FM) proposes a scheme whereby less measurements are collected during sensing and reconstruction is performed to recover the image. Much work has gone into optimizing the sensing and reconstruction portions separately. We propose a method of jointly optimizing both sensing and reconstruction end-to-end under a total measurement constraint, enabling learning of the optimal sensing scheme concurrently with the parameters of a neural network-based reconstruction network. We train our model on a rich dataset of confocal, two-photon, and wide-field microscopy images comprising of a variety of biological samples. We show that our method outperforms several baseline sensing schemes and a regularized regression reconstruction algorithm. Our code is publicly-available at https://github.com/alanqrwang/csfm.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Note we are not considering the effects of a point-spread function here.
- 2.
Note we are defining one measurement to be the computation of one Hadamard coefficient without averaging, despite the fact that this is a two-step process in the physical imaging setup.
- 3.
We used publicly-available code from: https://github.com/alanqrwang/HQSNet.
- 4.
Negative values of b were replaced with 0 following the paper’s experiments [33].
References
Bahadir, C.D., Wang, A.Q., Dalca, A.V., Sabuncu, M.R.: Deep-learning-based optimization of the under-sampling pattern in MRI. IEEE Trans. Comput. Imaging 6, 1139–1152 (2020)
Beck, A., Teboulle, M.: A fast iterative shrinkage-thresholding algorithm for linear inverse problems. SIAM J. Imaging Sci. 2(1), 183–202 (2009)
Boyd, S., Parikh, N., Chu, E., Peleato, B., Eckstein, J.: Distributed optimization and statistical learning via the alternating direction method of multipliers. Found. Trends Mach. Learn. 3(1), 1–122 (2011)
Chakrabarti, A.: Learning sensor multiplexing design through back-propagation. In: Proceedings of the 30th International Conference on Neural Information Processing Systems, NIPS 2016, pp. 3089–3097. Curran Associates Inc., Red Hook (2016)
Diamond, S., Sitzmann, V., Heide, F., Wetzstein, G.: Unrolled optimization with deep priors. CoRR abs/1705.08041 (2017)
Foi, A., Trimeche, M., Katkovnik, V., Egiazarian, K.: Practical poissonian-gaussian noise modeling and fitting for single-image raw-data. IEEE Trans. Image Process. 17(10), 1737–1754 (2008)
Gibson, G.M., Johnson, S.D., Padgett, M.J.: Single-pixel imaging 12 years on: a review. Opt. Express 28(19), 28190–28208 (2020)
Hopt, A., Neher, E.: Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys. J . 80(4), 2029–2036 (2001)
Jang, E., Gu, S., Poole, B.: Categorical reparametrization with gumble-softmax. In: Proceedings of the International Conference on Learning Representations 2017. OpenReviews.net, April 2017
Kellman, M., Bostan, E., Chen, M., Waller, L.: Data-driven design for Fourier ptychographic microscopy. In: 2019 IEEE International Conference on Computational Photography (ICCP), pp. 1–8 (2019)
Lee, S., Negishi, M., Urakubo, H., Kasai, H., Ishii, S.: Mu-net: multi-scale U-net for two-photon microscopy image denoising and restoration. Neural Netw. 125, 92–103 (2020)
Lichtman, J., Conchello, J.: Fluorescence microscopy. Nat. Methods 2, 910–919 (2005)
Maddison, C.J., Mnih, A., Teh, Y.W.: The concrete distribution: a continuous relaxation of discrete random variables. In: International Conference on Learning Representations (2017)
Magidson, V., Khodjakov, A.: Circumventing photodamage in live-cell microscopy. In: Sluder, G., Wolf, D.E. (eds.) Digital Microscopy, Methods in Cell Biology, vol. 114, pp. 545–560. Academic Press (2013)
Makitalo, M., Foi, A.: Optimal inversion of the generalized Anscombe transformation for Poisson-Gaussian noise. IEEE Trans. Image Process. 22(1), 91–103 (2013)
Parot, V.J., et al.: Compressed Hadamard microscopy for high-speed optically sectioned neuronal activity recordings. J. Phys. D Appl. Phys. 52(14), 144001 (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sitzmann, V., et al.: End-to-end optimization of optics and image processing for achromatic extended depth of field and super-resolution imaging. ACM Trans. Graph. 37(4), 1–13 (2018)
Streeter, L., Burling-Claridge, G.R., Cree, M.J., Künnemeyer, R.: Optical full Hadamard matrix multiplexing and noise effects. Appl. Opt. 48(11), 2078–2085 (2009)
Studer, V., Bobin, J., Chahid, M., Mousavi, H.S., Candes, E., Dahan, M.: Compressive fluorescence microscopy for biological and hyperspectral imaging. Proc. Natl. Acad. Sci. 109(26), E1679–E1687 (2012)
Sun, H., Dalca, A.V., Bouman, K.L.: Learning a probabilistic strategy for computational imaging sensor selection. In: 2020 IEEE International Conference on Computational Photography (ICCP), pp. 1–12 (2020)
Sun, M.J., Meng, L.T., Edgar, M.: A Russian Dolls ordering of the Hadamard basis for compressive single-pixel imaging. Sci. Rep. 7 (2017). Article number: 3464
Wang, A.Q., Dalca, A.V., Sabuncu, M.R.: Neural network-based reconstruction in compressed sensing MRI without fully-sampled training data. In: Deeba, F., Johnson, P., Würfl, T., Ye, J.C. (eds.) MLMIR 2020. LNCS, vol. 12450, pp. 27–37. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-61598-7_3
Wang, H., Rivenson, Y., Jin, Y.: Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019)
Wang, Z., Bovik, A.C., Sheikh, H.R., Simoncelli, E.P.: Image quality assessment: from error visibility to structural similarity. Trans. Image Process. 13(4), 600–612 (2004)
Weigert, M., Schmidt, U., Boothe, T.: Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15(12), 1090–1097 (2018)
Wijesinghe, P., Escobet-Montalbán, A., Chen, M., Munro, P.R.T., Dholakia, K.: Optimal compressive multiphoton imaging at depth using single-pixel detection. Opt. Lett. 44(20), 4981 (2019)
Xue, Y., Bigras, G., Hugh, J., Ray, N.: Training convolutional neural networks and compressed sensing end-to-end for microscopy cell detection. IEEE Trans. Med. Imaging 38(11), 2632–2641 (2019)
Yang, Y., Sun, J., Li, H., Xu, Z.: Deep ADMM-Net for compressive sensing MRI. In: Lee, D., Sugiyama, M., Luxburg, U., Guyon, I., Garnett, R. (eds.) Advances in Neural Information Processing Systems, vol. 29. Curran Associates, Inc. (2016)
Yao, R., Ochoa, M., Yan, P.: Net-FLICS: fast quantitative wide-field fluorescence lifetime imaging with compressed sensing - a deep learning approach. Light Sci. Appl. 8 (2019). Article number: 26
Yu, X., Yang, F., Gao, B., Ran, J., Huang, X.: Deep compressive single pixel imaging by reordering Hadamard basis: a comparative study. IEEE Access 8, 55773–55784 (2020)
Zhang, J., et al.: Extending LOUPE for K-space under-sampling pattern optimization in multi-coil MRI. In: Deeba, F., Johnson, P., Würfl, T., Ye, J.C. (eds.) MLMIR 2020. LNCS, vol. 12450, pp. 91–101. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-61598-7_9
Zhang, Y., et al.: A Poisson-Gaussian denoising dataset with real fluorescence microscopy images. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 11702–11710 (2019)
Acknowledgments
This work was, in part, supported by NIH R01 grants (R01LM012719 and R01AG053949, to MRS), the NSF NeuroNex grant (1707312, to MRS), an NSF CAREER grant (1748377, to MRS), and an NSF NeuroNex Hub grant (DBI-1707312, to CX).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, A.Q., LaViolette, A.K., Moon, L., Xu, C., Sabuncu, M.R. (2021). Joint Optimization of Hadamard Sensing and Reconstruction in Compressed Sensing Fluorescence Microscopy. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12906. Springer, Cham. https://doi.org/10.1007/978-3-030-87231-1_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-87231-1_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87230-4
Online ISBN: 978-3-030-87231-1
eBook Packages: Computer ScienceComputer Science (R0)