Abstract
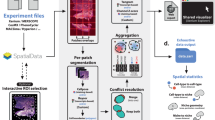
Spatial transcriptomics techniques such as STARmap [15] enable the subcellular detection of RNA transcripts within complex tissue sections. The data from these techniques are impacted by optical microscopy limitations, such as shading or vignetting effects from uneven illumination during image capture. Downstream analysis of these sparse spatially resolved transcripts is dependent upon the correction of these artefacts. This paper introduces a novel non-parametric vignetting correction tool for spatial transcriptomic images, which estimates the illumination field and background using an efficient iterative sliced histogram normalization routine. We show that our method outperforms the state-of-the-art shading correction techniques both in terms of illumination and background field estimation and requires fewer input images to perform the estimation adequately. We further demonstrate an important downstream application of our technique, showing that spatial transcriptomic volumes corrected by our method yield a higher and more uniform gene expression spot-calling in the rodent hippocampus. Python code and a demo file to reproduce our results are provided in the supplementary material and at this github page: https://github.com/BoveyRao/Non-parametric-vc-for-sparse-st.
B. Y. Rao and A. M. Peterson—These authors contributed equally.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chen, S., Loper, J., Chen, X., Zador, T., Paninski, L.: Barcode demixing through non-negative spatial regression (BarDensr). bioRxiv (2020)
Guizar-Sicairos, M., Thurman, S.T., Fienup, J.R.: Efficient subpixel image registration algorithms. Opt. Lett. 33(2), 156–158 (2008)
Kim, S.J., Pollefeys, M.: Robust radiometric calibration and vignetting correction. IEEE Trans. Pattern Anal. Mach. Intell. 30(4), 562–576 (2008)
Kolouri, S., Zou, Y., Rohde, G.K.: Sliced Wasserstein kernels for probability distributions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5258–5267 (2016)
Lee, J.K., Chung, J., Druey, K.M., Tansey, M.G.: Rgs10 exerts a neuroprotective role through the PKA/C-AMP response-element (CREB) pathway in dopaminergic neuron-like cells. J. Neurochem. 122(2), 333–343 (2012)
Lee, J.H., et al.: Highly multiplexed subcellular RNA sequencing in situ. Science 343(6177), 1360–1363 (2014)
Mitra, R.D., Shendure, J., Olejnik, J., Church, G.M., et al.: Fluorescent in situ sequencing on polymerase colonies. Anal. Biochem. 320(1), 55–65 (2003)
Oreopoulos, J., Berman, R., Browne, M.: Spinning-disk confocal microscopy: present technology and future trends. Methods Cell Biol. 123, 153–175 (2014)
Peng, T., et al.: A basic tool for background and shading correction of optical microscopy images. Nat. Commun. 8(1), 1–7 (2017)
Peng, T., Wang, L., Bayer, C., Conjeti, S., Baust, M., Navab, N.: Shading correction for whole slide image using low rank and sparse decomposition. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8673, pp. 33–40. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10404-1_5
Piccinini, F., Bevilacqua, A., Smith, K., Horvath, P.: Vignetting and photo-bleaching correction in automated fluorescence microscopy from an array of overlapping images. In: 2013 IEEE 10th International Symposium on Biomedical Imaging, pp. 464–467. IEEE (2013)
Smith, K., et al.: CIDRE: an illumination-correction method for optical microscopy. Nat. Methods 12(5), 404–406 (2015)
Stosiek, C., Garaschuk, O., Holthoff, K., Konnerth, A.: In vivo two-photon calcium imaging of neuronal networks. Proc. Natl. Acad. Sci. 100(12), 7319–7324 (2003)
Tomaževič, D., Likar, B., Pernuš, F.: Comparative evaluation of retrospective shading correction methods. J. Microsc. 208(3), 212–223 (2002)
Wang, X., et al.: Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361(6400) (2018)
Zeisel, A., et al.: Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-Seq. Science 347(6226), 1138–1142 (2015)
Zheng, Y., Lin, S., Kambhamettu, C., Yu, J., Kang, S.B.: Single-image vignetting correction. IEEE Trans. Pattern Anal. Mach. Intell. 31(12), 2243–2256 (2008)
Acknowledgements
BYR, SH, and AL are supported by NIMH 1R01MH124047 and 1R01MH124867; and NINDS 1U19NS104590 and 1U01NS115530. SH also is supported by NIMH 5K00MH121382. AMP, EKK, and AHR are funded from CZF2019-002460 and 1R01MH124047-01. LP, EV are supported by Simons Foundation 543023, NSF 1912194, NSF NeuroNex Award 1707398 and The Gatsby Charitable Foundation GAT3708.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Rao, B.Y. et al. (2021). Non-parametric Vignetting Correction for Sparse Spatial Transcriptomics Images. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12908. Springer, Cham. https://doi.org/10.1007/978-3-030-87237-3_45
Download citation
DOI: https://doi.org/10.1007/978-3-030-87237-3_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87236-6
Online ISBN: 978-3-030-87237-3
eBook Packages: Computer ScienceComputer Science (R0)