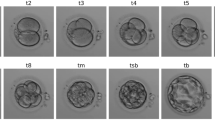
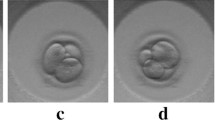
Abstract
In Vitro Fertilization (IVF) treatment is increasingly chosen by couples suffering from infertility as a means to conceive. Time-lapse imaging technology has enabled continuous monitoring of embryos in vitro and time-based development metrics for assessing embryo quality prior to transfer. Timing at which embryos reach certain development stages provides valuable information about their potential to become a positive pregnancy. Automating development stage detection of day 4–5 embryos remains difficult due to small variation between stages. In this paper, a classifier is trained to detect embryo development stage with learning strategies added to explicitly address challenges of this task. Synergic loss encourages the network to recognize and utilize stage similarities between different embryos. Short-range temporal learning incorporates chronological order to embryo sequence predictions. Image and sequence augmentations complement both approaches to increase generalization to unseen sequences. The proposed approach was applied to human embryo sequences with labeled morula and blastocyst stage onsets. Morula and blastocyst stage classification was improved by 5.71% and 1.11%, respectively, while morula and blastocyst stage mean absolute onset error was reduced by 19.1% and 8.7%, respectively. Code is available: https://github.com/llockhar/Embryo-Stage-Onset-Detection.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
National Center for Chronic Disease Prevention and Health Promotion: Division of Reproductive Health: 2018 Assisted Reproductive Technology National Summary Report. https://www.cdc.gov/art/reports/2018/fertility-clinic.html. Accessed 9 Feb 2021
Rienzi, L., et al.: Time of morulation and trophectoderm quality are predictors of a live birth after euploid blastocyst transfer: a multicenter study. Fertil. Steril. 112(6), 1080–1093 (2019)
Motato, Y., de los Santos, M.J., Escriba, M.J., Ruiz, B.A., Remohí, J., Meseguer, M.: Morphokinetic analysis and embryonic prediction for blastocyst formation through an integrated time-lapse system. Fertil. Steril. 105(2), 376–384 (2016)
Desai, N., Ploskonka, S., Goodman, L.R., Austin, C., Goldberg, J., Falcone, T.: Analysis of embryo morphokinetics, multinucleation and cleavage anomalies using continuous time-lapse monitoring in blastocyst transfer cycles. Reprod. Biol. Endocrinol. 12(1), 1–10 (2014)
Jacobs, C., et al.: Correlation between morphokinetic parameters and standard morphological assessment: what can we predict from early embryo development? a time-lapse-based experiment with 2085 blastocysts. JBRA Assist. Reprod. 24(3), 273 (2020)
Basile, N., et al.: The use of morphokinetics as a predictor of implantation: A multicentric study to define and validate an algorithm for embryo selection. Hum. Reprod. 30(2), 276–283 (2014)
Meseguer, M., Herrero, J., Tejera, A., Hilligsøe, K.M., Ramsing, N.B., Remohí, J.: The use of morphokinetics as a predictor of embryo implantation. Hum. Reprod. 26(10), 2658–2671 (2011)
Cruz, M., Garrido, N., Herrero, J., Pérez-Cano, I., Muñoz, M., Meseguer, M.: Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod. Biomed. Online 25(4), 371–381 (2012)
Feyeux, M., et al.: Development of automated annotation software for human embryo morphokinetics. Hum. Reprod. 35(3), 557–564 (2020)
Leahy, B.D., et al.: Automated measurements of key morphological features of human embryos for IVF. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12265, pp. 25–35. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59722-1_3
Khan, A., Gould, S., Salzmann, M.: Deep convolutional neural networks for human embryonic cell counting. In: Hua, G., Jégou, H. (eds.) ECCV 2016. LNCS, vol. 9913, pp. 339–348. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46604-0_25
Ng, N.H., McAuley, J.J., Gingold, J., Desai, N., Lipton, Z.C.: Predicting embryo morphokinetics in videos with late fusion nets & dynamic decoders. In: International Conference on Learning Representations (Workshop) (2018)
Liu, Z., et al.: Multi-task deep learning with dynamic programming for embryo early development stage classification from time-lapse videos. IEEE Access 7, 122153–122163 (2019)
Dirvanauskas, D., Maskeliunas, R., Raudonis, V., Damasevicius, R.: Embryo development stage prediction algorithm for automated time lapse incubators. Comput. Methods Programs Biomed. 177, 161–174 (2019)
Raudonis, V., Paulauskaite-Taraseviciene, A., Sutiene, K., Jonaitis, D.: Towards the automation of early-stage human embryo development detection. Biomed. Eng. Online 18(1), 1–20 (2019)
Lau, T., Ng, N., Gingold, J., Desai, N., McAuley, J. and Lipton, Z.C.: Embryo staging with weakly-supervised region selection and dynamically-decoded predictions. In: Machine Learning for Healthcare Conference, pp. 663–679. PMLR (2019)
Malmsten, J., Zaninovic, N., Zhan, Q., Rosenwaks, Z., Shan, J.: Automated cell division classification in early mouse and human embryos using convolutional neural networks. Neural Comput. Appl. 33(7), 2217–2228 (2020). https://doi.org/10.1007/s00521-020-05127-8
Hochreiter, S., Schmidhuber, J.: Long short-term memory. Neural Comput. 7, 1735–1780 (1997)
Simonyan, K. and Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, (2014)
Zhang, J., Xie, Y., Wu, Q., Xia, Y.: Medical image classification using synergic deep learning. Med. Image Anal. 54, 10–19 (2019)
Zhang, H., Cisse, M., Dauphin, Y.N., Lopez-Paz, D.: mixup: Beyond empirical risk minimization. In: International Conference on Learning Representations (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Lockhart, L., Saeedi, P., Au, J., Havelock, J. (2021). Automating Embryo Development Stage Detection in Time-Lapse Imaging with Synergic Loss and Temporal Learning. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12905. Springer, Cham. https://doi.org/10.1007/978-3-030-87240-3_52
Download citation
DOI: https://doi.org/10.1007/978-3-030-87240-3_52
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87239-7
Online ISBN: 978-3-030-87240-3
eBook Packages: Computer ScienceComputer Science (R0)