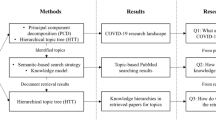
Abstract
The COVID-19 pandemic triggered a wave of novel scientific literature that is impossible to inspect and study in a reasonable time frame manually. Current machine learning methods offer to project such body of literature into the vector space, where similar documents are located close to each other, offering an insightful exploration of scientific papers and other knowledge sources associated with COVID-19. However, to start searching, such texts need to be appropriately annotated, which is seldom the case due to the lack of human resources. In our system, the current body of COVID-19-related literature is annotated using unsupervised keyphrase extraction, facilitating the initial queries to the latent space containing the learned document embeddings (low-dimensional representations). The solution is accessible through a web server capable of interactive search, term ranking, and exploration of potentially interesting literature. We demonstrate the usefulness of the approach via case studies from the medicinal chemistry domain.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
- 9.
- 10.
- 11.
- 12.
- 13.
- 14.
- 15.
- 16.
References
The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5(4), 536–544 (2020). https://doi.org/10.1038/s41564-020-0695-z
Advani, I., et al.: Is increased sleep responsible for reductions in myocardial infarction during the COVID-19 pandemic? Am. J. Cardiol. 131, 128–130 (2020)
Agarwal, S., Kaushik, J.S.: Student’s perception of online learning during COVID pandemic. Indian J. Pediatr. 87(7), 554 (2020). https://doi.org/10.1007/s12098-020-03327-7
Buonaguro, L., Buonaguro, F.M.: Knowledge-based repositioning of the anti-HCV direct antiviral agent sofosbuvir as SARS-CoV-2 treatment. Infect. Agents Cancer 15(1) (2020). https://doi.org/10.1186/s13027-020-00302-x
Campos, R., Mangaravite, V., Pasquali, A., Jorge, A., Nunes, C., Jatowt, A.: YAKE! Keyword extraction from single documents using multiple local features. Inf. Sci. 509, 257–289 (2020)
Cattaneo, C.: Forensic medicine in the time of COVID 19: an editorial from Milano, Italy. Forensic Sci. Int. 312, 110308 (2020)
Chew, C., Eysenbach, G.: Pandemics in the age of Twitter: content analysis of tweets during the 2009 H1N1 outbreak. PLoS ONE 5(11), e14118 (2010)
El-Kassas, W.S., Salama, C.R., Rafea, A.A., Mohamed, H.K.: Automatic text summarization: a comprehensive survey. Expert Syst. Appl. 165, 113679 (2021)
Fani, M., Teimoori, A., Ghafari, S.: Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virol. 15(5), 317–323 (2020)
Gates, B.: Responding to COVID-19 – a once-in-a-century pandemic? N. Engl. J. Med. 382(18), 1677–1679 (2020)
Hasan, K.S., Ng, V.: Automatic keyphrase extraction: a survey of the state of the art. In: Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pp. 1262–1273 (2014)
Hing, C., Al-Dadah, O.: Returning to elective surgery, the ‘new normal’. Knee 27(3), A1 (2020)
Honore, P.M., et al.: Therapeutic plasma exchange as a routine therapy in septic shock and as an experimental treatment for COVID-19: we are not sure. Critical Care 24(1) (2020). https://doi.org/10.1186/s13054-020-02943-1
Hutson, M.: Artificial-intelligence tools aim to tame the coronavirus literature. Nature (2020). https://www.nature.com/articles/d41586-020-01733-7
Ijaz, M.K., et al.: Microbicidal actives with virucidal efficacy against SARS-CoV-2. Am. J. Infect. Control 48(8), 972–973 (2020)
Jin, Z., et al.: Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582(7811), 289–293 (2020)
Jones, S., Lundy, S., Paynter, G.W.: Interactive document summarisation using automatically extracted keyphrases. In: Proceedings of the 35th Annual Hawaii International Conference on System Sciences, pp. 1160–1169. IEEE (2002)
Kilbourne, E.D.: Influenza pandemics of the 20th century. Emerg. Infect. Dis. 12(1), 9–14 (2006)
Kumar, S., Nyodu, R., Maurya, V.K., Saxena, S.K.: Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In: Saxena, S.K. (ed.) Coronavirus Disease 2019 (COVID-19). MVFPDC, pp. 23–31. Springer, Singapore (2020). https://doi.org/10.1007/978-981-15-4814-7_3
Le, Q., Mikolov, T.: Distributed representations of sentences and documents. In: Proceedings of the 31st International Conference on International Conference on Machine Learning - Volume 32, ICML 2014, pp. II-1188–II-1196. JMLR.org (2014)
Le Bras, P., Gharavi, A., Robb, D., Vidal, A., Padilla, S., Chantler, M.: Visualising COVID-19 research. Working paper, arXiv, May 2020
Li, H., Zhou, Y., Zhang, M., Wang, H., Zhao, Q., Liu, J.: Updated approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 64(6) (2020). https://doi.org/10.1128/aac.00483-20
Lutchman, D.: Could the smoking gun in the fight against COVID-19 be the (rh)ACE-2? Eur. Respir. J. 56(1), 2001560 (2020)
Matsuyama, S., et al.: Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. 117(13), 7001–7003 (2020)
McInnes, L., Healy, J., Saul, N., Großberger, L.: UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3(29), 861 (2018). https://doi.org/10.21105/joss.00861
Mohseni, A.H., Taghinezhad-S, S., Xu, Z., Fu, X.: Body fluids may contribute to human-to-human transmission of severe acute respiratory syndrome coronavirus 2: evidence and practical experience. Chin. Med. 15(1) (2020). https://doi.org/10.1186/s13020-020-00337-7
Novins, D.K., et al.: JAACAP’s role in advancing the science of pediatric mental health and promoting the care of youth and families during the COVID-19 pandemic. J. Am. Acad. Child Adolesc. Psychiatry 59(6), 686–688 (2020)
Ortega, J.T., Serrano, M.L., Pujol, F.H., Rangel, H.R.: Role of changes in SARS-COV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 19, Doc410 (2020). https://doi.org/10.17179/EXCLI2020-1167. ISSN 1611–2156, https://www.excli.de/vol19/Rangel_18032020_proof.pdf
Panciani, P.P., et al.: SARS-CoV-2: “three-steps’’ infection model and CSF diagnostic implication. Brain Behav. Immunity 87, 128–129 (2020)
Randolph, G.W.: One virus, undivided ... equity, and the corona virus. Laryngoscope Investigative Otolaryngol. 5(3), 586–589 (2020). https://doi.org/10.1002/lio2.398
Saxena, S.K., Kumar, S., Maurya, V.K., Sharma, R., Dandu, H.R., Bhatt, M.L.B.: Current insight into the novel coronavirus disease 2019 (COVID-19). In: Saxena, S.K. (ed.) Coronavirus Disease 2019 (COVID-19). MVFPDC, pp. 1–8. Springer, Singapore (2020). https://doi.org/10.1007/978-981-15-4814-7_1
Škrlj, B., Repar, A., Pollak, S.: RaKUn: Rank-based Keyword extraction via Unsupervised learning and meta vertex aggregation. In: Martín-Vide, C., Purver, M., Pollak, S. (eds.) SLSP 2019. LNCS (LNAI), vol. 11816, pp. 311–323. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-31372-2_26
Su, S., Jiang, S.: A suspicious role of interferon in the pathogenesis of SARS-CoV-2 by enhancing expression of ACE2. Signal Transduction Targeted Therapy 5(1) (2020). https://doi.org/10.1038/s41392-020-0185-z
Tiwari, V., Beer, J.C., Sankaranarayanan, N.V., Swanson-Mungerson, M., Desai, U.R.: Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today 25(8), 1535–1544 (2020)
Wang, C., Horby, P.W., Hayden, F.G., Gao, G.F.: A novel coronavirus outbreak of global health concern. Lancet 395(10223), 470–473 (2020)
Wang, D., et al.: Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323(11), 1061 (2020). https://doi.org/10.1001/jama.2020.1585
Wang, L.L., Lo, K.: Text mining approaches for dealing with the rapidly expanding literature on COVID-19. Brief. Bioinform. 22(2), 781–799 (2020). https://doi.org/10.1093/bib/bbaa296
Wang, L.L., et al.: CORD-19: the COVID-19 open research dataset. arXiv (2020)
Whitacre, R.P., Buchbinder, L.S., Holmes, S.M.: The pandemic present. Soc. Anthropol. 28(2), 380–382 (2020)
Wu, C., et al.: Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B 10(5), 766–788 (2020)
Zhang, H., Penninger, J.M., Li, Y., Zhong, N., Slutsky, A.S.: Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46(4), 586–590 (2020)
Zhou, H., Fang, Y., Xu, T., Ni, W.J., Shen, A.Z., Meng, X.M.: Potential therapeutic targets and promising drugs for combating SARS-CoV-2. Br. J. Pharmacol. 177(14), 3147–3161 (2020)
Acknowledgements
This work was supported by the Slovenian Research Agency (ARRS) core research program P2-0103 and the CRP project V3-2033. The work of the first author was financed by the ARRS young researchers grant. The work was also supported by European Union’s Horizon 2020 research and innovation programme under grant agreement No 825153, project EMBEDDIA (Cross-Lingual Embeddings for Less-Represented Languages in European News Media).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Škrlj, B., Jukič, M., Eržen, N., Pollak, S., Lavrač, N. (2021). Prioritization of COVID-19-Related Literature via Unsupervised Keyphrase Extraction and Document Representation Learning. In: Soares, C., Torgo, L. (eds) Discovery Science. DS 2021. Lecture Notes in Computer Science(), vol 12986. Springer, Cham. https://doi.org/10.1007/978-3-030-88942-5_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-88942-5_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-88941-8
Online ISBN: 978-3-030-88942-5
eBook Packages: Computer ScienceComputer Science (R0)