Abstract
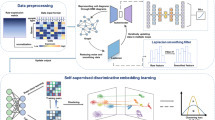
Single-cell RNA-seq (scRNA-seq) data has provided a higher resolution of cellular heterogeneity. However, scRNA-seq data also brings some computational challenges for its high-dimension, high-noise, and high-sparseness. The dimension reduction is a crucial way to denoise and greatly reduce the computational complexity by representing the original data in a low-dimensional space. In this study, to achieve an accurate low-dimension representation, we proposed a denoising AutoEncoder based dimensionality reduction method for scRNA-seq data (ScDA), combining the denoising function with the AutoEncoder. ScDA is a deep unsupervised generative model, which models the dropout events and denoises the scRNA-seq data. Meanwhile, ScDA can reveal the nonlinear feature extraction of the original data through maximum distribution similarity before and after dimensionality reduction. Tested on 16 scRNA-seq datasets, ScDA provides superior average performances, and especially superior performances in large-scale datasets compared with 3 clustering methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Authors’ Contributions
Conceptualization, X.Z., X.P., and J.W.; Methodology, X.Z., Y.L., and X.P.; Software, Y.L., and J.L.; Writing-Original Draft Preparation, X.Z., J.W., and X.P.; Visualization, Y.L.; Funding Acquisition, X.Z., and X.P.
References
Vitak, S.A., et al.: Sequencing thousands of single-cell genomes with combinatorial indexing. Nat. Methods 14(3), 302 (2017)
Stuart, T., Satija, R.: Integrative single-cell analysis. Nat. Rev. Genet. 20(5), 257–272 (2019)
Laehnemann, D., Kster, J., Szczurek, E., Mccarthy, D.J., Schnhuth, A.: Eleven grand challenges in single-cell data science. Genome Biol. 21(1), 31 (2020)
Wolf, F.A., et al.: PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20(1), 59 (2019)
Taiyun, K., Chen, I.R., Lin, Y., Wang, Y.Y., Yang, J., Yang, P.: Impact of similarity metrics on single-cell RNA-seq data clustering. Brief. Bioinform. 20(6), 2316–2326 (2018)
Eling, N., Morgan, M.D., Marioni, J.C.: Challenges in measuring and understanding biological noise. Nat. Rev. Genet. 20(9), 536–548 (2019)
Andrews, T.S., Hemberg, M., Birol, I.: Dropout-based feature selection for scRNASeq. Bioinformatics 35(16), 2865–2867 (2018)
Wang, D.: VASC: Dimension reduction and visualization of single-cell RNA-seq data by deep variational autoencoder. Genomics Proteomics Bioinformatics 16(5), 320–331 (2018)
Raphael, P., Li, Z., Kuang, R.: Machine learning and statistical methods for clustering single-cell RNA-sequencing data. Brief. Bioinform. 4, 4 (2019)
Kiselev, V.Y., et al.: SC3: consensus clustering of single-cell RNA-seq data. Nat. Methods 14(5), 483 (2017)
Jia, C., Hu, Y., Derek, K., Junhyong, K., Li, M., Zhang, N.R.: Accounting for technical noise in differential expression analysis of single-cell RNA sequencing data. Nucleic Acids Res. 19, 10978 (2017)
Liu, Z., et al.: Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nat. Commun. 8(1), 22 (2017)
Saelens, W., Cannoodt, R., Todorov, H., Saeys, Y.: A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37(5), 547–554 (2019)
Llorens-Bobadilla, E., Zhao, S., Baser, A., Saiz-Castro, G., Zwadlo, K., Martin-Villalba, A.: Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17(3), 329–340 (2015)
Spyros, D., et al.: Hayden, Barres BA, Quake SR: A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. 112(23), 7285–7290 (2015)
GiniClust3: a fast and memory-efficient tool for rare cell type identification. BMC Bioinformatics 21(1), 158 (2020)
Zhu, X., Zhang, J., Xu, Y., Wang, J., Peng, X., Li, H.-D.: Single-cell clustering based on shared nearest neighbor and graph partitioning. Interdisc. Sci.: Computat. Life Sci. (2020)
Yip, S.H., Chung, S.P., Wang, J.: Evaluation of tools for highly variable gene discovery from single-cell RNA-seq data. Brief. Bioinform. 4, 4 (2018)
Becht, E., et al.: Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019)
Tian, T., Wan, J., Song, Q., Wei, Z.: Clustering single-cell RNA-seq data with a model-based deep learning approach. Nat. Mach. Intell. 1(4), 191–198 (2019)
Wan, S., Kim, J., Won, K.J.: SHARP: hyper-fast and accurate processing of single-cell RNA-seq data via ensemble random projection. Genome Res. 30(2), gr.254557.254119 (2020)
Liang, Z., Li, M., Zheng, R., Tian, Y., Wang, J.: SSRE: cell type detection based on sparse subspace representation and similarity enhancement. Genomics Proteomics Bioinformatics S1762–0229(21), 00038–33 (2020)
Song, J., Liu, Y., Zhang, X., Wu, Q., Yang, C.: Entropy subspace separation-based clustering for noise reduction (ENCORE) of scRNA-seq data. Nucleic Acids Res. 49(3), e18 (2020)`
Kiselev, V.Y., Andrews, T.S., Hemberg, M.: Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20(5), 273–282 (2019)
Soneson, C., Robinson, M.D.: Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods 15(4), 255–261 (2018)
Estevez, P.A., Tesmer, M., Perez, C.A., Zurada, J.M.: Normalized mutual information feature selection. IEEE Trans. Neural Netw. 20(2), 189–201 (2009)
Hubert, L., Arabie, P.: Comparing partitions. J. Classif. 2(1), 193–218 (1985)
Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165(1), 61–74 (2016)
Kumar, R.M., et al.: Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 16(7529), 56–61 (2014)
Wang, Y.J., et al.: Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65(10), 3028 (2016)
Wallrapp, A., et al.: The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549(7672), 351–356 (2017)
Patel, A.P., et al.: Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344(6190), 1396–1401 (2014)
Haber, A.L., et al.: A single-cell survey of the small intestinal epithelium. Nature 551(7680), 333–339 (2017)
Petropoulos, S., et al.: Single-cell RNA-Seq reveals lineage and x chromosome dynamics in human preimplantation embryos. Cell 165(4), 1012–1026 (2016)
Klein, A., et al.: Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161(5), 1187–1201 (2015)
Han, X., Wang, R., Zhou, Y., Fei, L., Guo, G.: Mapping the mouse cell atlas by Microwell-Seq. Cell 172(5), 1091-1107.e1017 (2018)
Grün, D., et al.: De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19(2), 266–277 (2016)
Cao, J., et al.: Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357(6352), 661 (2017)
Spallanzani, R.G., Zemmour, D., Xiao, T., Jayewickreme, T., Mathis, D.: Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol. 4(35), eaaw3658 (2019)
Zemmour, D., Zilionis, R., Kiner, E., Klein, A.M., Mathis, D., Benoist, C.: Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19(3), 291–301 (2018)
Frigerio, C.S., et al.: The major risk factors for Alzheimer’s disease: age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep. 27(4), 1293-1306.e1296 (2019)
Shekhar, K., et al.: Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166(5), 1308-1323.e1330 (2016)
Funding
This research was supported by the National Natural Science Foundation of China (No. 61762087, 61702555, 61772557), Hunan Provincial Science and Technology Program (2018WK4001), 111 Project (No. B18059), Guangxi Natural Science Foundation (No. 2018JJA170175).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhu, X., Lin, Y., Li, J., Wang, J., Peng, X. (2021). ScDA: A Denoising AutoEncoder Based Dimensionality Reduction for Single-cell RNA-seq Data. In: Wei, Y., Li, M., Skums, P., Cai, Z. (eds) Bioinformatics Research and Applications. ISBRA 2021. Lecture Notes in Computer Science(), vol 13064. Springer, Cham. https://doi.org/10.1007/978-3-030-91415-8_45
Download citation
DOI: https://doi.org/10.1007/978-3-030-91415-8_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-91414-1
Online ISBN: 978-3-030-91415-8
eBook Packages: Computer ScienceComputer Science (R0)