Abstract
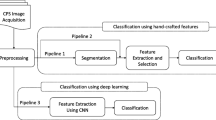
Morphological changes in the cell structure in Pap Smear images are the basis for classification in pathology. Identification of this classification is a challenge because of the complexity of Pap Smear images caused by changes in cell morphology. This procedure is very important because it provides basic information for detecting cancerous or precancerous lesions. To help advance research in this area, we present the RepoMedUNM Pap smear image database consisting of non-ThinPrep (nTP) Pap test images and ThinPrep (TP) Pap test images. It is common for research groups to have their image datasets. This need is driven by the fact that established datasets are not publicly accessible. The purpose of this study is to present the RepoMedUNM dataset analysis performed for texture feature cells on new images consisting of four classes, normal, L-Sil, H-Sil, and Koilocyt with K-means segmentation. Evaluation of model classification using reuse pretrained network method. Convolutional Neural Network (CNN) implements the pre-trained CNN VGG16, VGG19, and ResNet50 models for the classification of three groups namely TP, nTP and all datasets. The results of feature cells and classification can be used as a reference for the evaluation of future classification techniques.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Plissiti, M.E., Dimitrakopoulos, P., Sfikas, G., Nikou, C., Krikoni, O., Charchanti, A.: SIPAKMED: a new dataset for feature and image based classification of normal and pathological cervical cells in Pap smear images. In: 2018 25th IEEE International Conference on Image Processing (ICIP), pp. 3144–3148. IEEE, October 2018
Riana, D., Plissiti, M.E., Nikou, C., Widyantoro, D.H., Mengko, T.L.R., Kalsoem, O.: Inflammatory cell extraction and nuclei detection in Pap smear images. Int. J. E-Health Med. Commun. (IJEHMC) 6(2), 27–43 (2015)
Kiran G.V.K., Reddy, G.M.: Automatic classification of whole slide pap smear images using CNN With PCA based feature interpretation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (2019)
Khamparia, A., Gupta, D., de Albuquerque, V.H.C., Sangaiah, A.K., Jhaveri, R.H.: Internet of health things-driven deep learning system for detection and classification of cervical cells using transfer learning. J. Supercomput. 76(11), 8590–8608 (2020). https://doi.org/10.1007/s11227-020-03159-4
Chen, Y.F., et al.: Semi-automatic segmentation and classification of pap smear cells. IEEE J. Biomed. Heal. Informat. 18, 94–108 (2014). https://doi.org/10.1109/JBHI.2013.2250984
Human Papillomavirus (HPV) and Cervical Cancer. Accessed July 2021. https://www.who.int/news-room/fact-sheets/detail/human-papillomavirus-(hpv)-and-cervical-cancer
Jantzen, J., Norup, J., Dounias, G., Bjerregaard, B.: Pap-smear benchmark data for pattern classification. In: Proceedings of Nature inspired Smart Information Systems (NiSIS), pp. 1–9 (2005)
Riana, D., et al.: Repository Medical Imaging Citra Pap Smear. http://repomed.nusamandiri.ac.id/ (2021)
Walker, R.F., Jackway, P., Lovell, B., Longstaff, I.D.: Classification of Cevical cell nuclei using morphological segmentation and textural feature extraction. In: Proceedings of ANZIIS 1994-Australian New Zealnd Intelligent Information Systems Conference, pp. 297–301 (1994)
Sulaiman, S.N., Isa, N.A.M., Yusoff, I.A., Othman, N.H.: Overlapping cells separation method for cervical cell images. In: Proceedings of the 2010 10th International Conference on Intelligence System Design and Applications ISDA-10 1218–1222 (2010). https://doi.org/10.1109/ISDA.2010.5687020
Merlina, N., Noersasongko, E., Nurtantio, P., Soeleman, M.A., Riana, D., Hadianti, S.: Detecting the width of pap smear cytoplasm image based on GLCM feature. In: Smart Trends in Computing and Communications: Proceedings of SmartCom 2020, pp. 231–239. Springer, Singapore (2021). https://doi.org/10.1007/978-981-15-5224-3_22
Riana, D., Rahayu, S., Hasan, M.: Comparison of segmentation and identification of Swietenia Mahagoni wood defects with augmentation images. Heliyon, e07417 (2021)
Liu, Z., et al.: Improved kiwifruit detection using pre-trained VGG16 with RGB and NIR information fusion. IEEE Access 8, 2327–2336 (2020). https://doi.org/10.1109/ACCESS.2019.2962513
Hridayami, P., Putra, I.K.G.D., Wibawa, K.S.: Fish species recognition using VGG16 deep convolutional neural network. J. Comput. Sci. Eng. 13(3), 124–130 (2019). https://doi.org/10.5626/JCSE.2019.13.3.124
Train Deep Learning Network to Classify New Images. Accessed 12 July 2021. Available: https://www.mathworks.com/help/deeplearning/ug/train-deep-learning-network-to-classify-new-images.html
BVLC GoogLeNet Model. Accessed 12 July 2021. Available: https://github.com/BVLC/caffe/tree/master/models/bvlc_googlenet
Lan, K., Wang, D.T., Fong, S., Liu, L.S., Wong, K.K., Dey, N.: A survey of data mining and deep learning in bioinformatics. J. Med. Syst. 42(8), 139 (2018)
Acknowledgments
The authors would like to thank the Directorate General of Higher Education, Ministry of Education, Culture, Research, and Technology Republik Indonesia for supporting this research through the national competitive applied research grant 2021.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Riana, D. et al. (2021). RepoMedUNM: A New Dataset for Feature Extraction and Training of Deep Learning Network for Classification of Pap Smear Images. In: Mantoro, T., Lee, M., Ayu, M.A., Wong, K.W., Hidayanto, A.N. (eds) Neural Information Processing. ICONIP 2021. Communications in Computer and Information Science, vol 1516. Springer, Cham. https://doi.org/10.1007/978-3-030-92307-5_37
Download citation
DOI: https://doi.org/10.1007/978-3-030-92307-5_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-92306-8
Online ISBN: 978-3-030-92307-5
eBook Packages: Computer ScienceComputer Science (R0)