Abstract
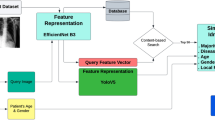
Chest radiography is one of the most common medical imaging modalites. However, chest radiography interpretation is a complex task that requires significant expertise. As such, the development of automatic systems for pathology detection has been proposed in literature, particularly using deep learning. However, these techniques suffer from a lack of explainability, which hinders their adoption in clinical scenarios. One technique commonly used by radiologists to support and explain decisions is to search for cases with similar findings for direct comparison. However, this process is extremely time-consuming and can be prone to confirmation bias. Automatic image retrieval methods have been proposed in literature but typically extract features from the whole image, failing to focus on the lesion in which the radiologist is interested. In order to overcome these issues, a novel framework LXIR for lesion-based image retrieval is proposed in this study, based on a state of the art object detection framework (YOLOv5) for the detection of relevant lesions as well as feature representation of those lesions. It is shown that the proposed method can successfully identify lesions and extract features which accurately describe high-order characteristics of each lesion, allowing to retrieve lesions of the same pathological class. Furthermore, it is show that in comparison to SSIM-based retrieval, a classical perceptual metric, and random retrieval of lesions, the proposed method retrieves the most relevant lesions 81% of times, according to the evaluation of two independent radiologists, in comparison to 42% of times by SSIM.
This work was funded by the ERDF - European Regional Development Fund, through the Programa Operacional Regional do Norte (NORTE 2020) and by National Funds through the FCT - Portuguese Foundation for Science and Technology, I.P. within the scope of the CMU Portugal Program (NORTE-01-0247-FEDER-045905) and UIDB/50014/2020.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bansal, G.: Digital radiography. A comparison with modern conventional imaging. Postgrad. Med. J. 82(969), 425–428 (2006)
Bay, H., Ess, A., Tuytelaars, T., Van Gool, L.: Speeded-up robust features (SURF). Comput. Vis. Image Underst. 110(3), 346–359 (2008)
Chen, B., Li, J., Guo, X., Lu, G.: DualCheXNet: dual asymmetric feature learning for thoracic disease classification in chest X-rays. Biomed. Signal Process. Control 53, 101554 (2019)
Cowan, I.A., MacDonald, S.L., Floyd, R.A.: Measuring and managing radiologist workload: measuring radiologist reporting times using data from a Radiology Information System. J. Med. Imaging Radiat. Oncol. 57(5), 558–566 (2013)
Gündel, S., Grbic, S., Georgescu, B., Liu, S., Maier, A., Comaniciu, D.: Learning to recognize abnormalities in chest X-rays with location-aware dense networks. In: Vera-Rodriguez, R., Fierrez, J., Morales, A. (eds.) CIARP 2018. LNCS, vol. 11401, pp. 757–765. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-13469-3_88
Hofmanninger, J., Langs, G.: Mapping visual features to semantic profiles for retrieval in medical imaging. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 457–465 (2015)
Irvin, J., et al.: CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 590–597 (2019)
Jocher, G., et al.: Ultralytics/YOLOv5 (2021). https://github.com/ultralytics/yolov5
Kumar, P., Grewal, M., Srivastava, M.M.: Boosted cascaded convnets for multilabel classification of thoracic diseases in chest radiographs. In: Campilho, A., Karray, F., ter Haar Romeny, B. (eds.) ICIAR 2018. LNCS, vol. 10882, pp. 546–552. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-93000-8_62
Li, Z., et al.: Thoracic disease identification and localization with limited supervision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8290–8299 (2018)
Li, Z., Zhang, X., Müller, H., Zhang, S.: Large-scale retrieval for medical image analytics: a comprehensive review. Med. Image Anal. 43, 66–84 (2018)
Lin, T.-Y., et al.: Microsoft COCO: common objects in context. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8693, pp. 740–755. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10602-1_48
Lowe, D.G.: Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vision 60(2), 91–110 (2004)
Nguyen, H.Q., et al.: VinDr-CXR: An open dataset of chest X-rays with radiologist’s annotations. arXiv preprint arXiv:2012.15029 (2020)
Redmon, J., Farhadi, A.: Yolov3: an incremental improvement. arXiv preprint arXiv:1804.02767 (2018)
Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D.: Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017)
Shin, H.C., Orton, M.R., Collins, D.J., Doran, S.J., Leach, M.O.: Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4d patient data. IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1930–1943 (2012)
Silva, W., Poellinger, A., Cardoso, J.S., Reyes, M.: Interpretability-guided content-based medical image retrieval. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12261, pp. 305–314. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59710-8_30
Wang, C.Y., Liao, H.Y.M., Wu, Y.H., Chen, P.Y., Hsieh, J.W., Yeh, I.H.: CSPNet: a new backbone that can enhance learning capability of CNN. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 390–391 (2020)
Wang, K., Liew, J.H., Zou, Y., Zhou, D., Feng, J.: PANet: few-shot image semantic segmentation with prototype alignment. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 9197–9206 (2019)
Wang, Z., Bovik, A.C., Sheikh, H.R., Simoncelli, E.P.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13(4), 600–612 (2004)
Wolterink, J.M., Leiner, T., Viergever, M.A., Išgum, I.: Automatic coronary calcium scoring in cardiac CT angiography using convolutional neural networks. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 589–596. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24553-9_72
Zhang, R., Isola, P., Efros, A.A., Shechtman, E., Wang, O.: The unreasonable effectiveness of deep features as a perceptual metric. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 586–595 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Pedrosa, J., Sousa, P., Silva, J., Mendonça, A.M., Campilho, A. (2022). Lesion-Based Chest Radiography Image Retrieval for Explainability in Pathology Detection. In: Pinho, A.J., Georgieva, P., Teixeira, L.F., Sánchez, J.A. (eds) Pattern Recognition and Image Analysis. IbPRIA 2022. Lecture Notes in Computer Science, vol 13256. Springer, Cham. https://doi.org/10.1007/978-3-031-04881-4_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-04881-4_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-04880-7
Online ISBN: 978-3-031-04881-4
eBook Packages: Computer ScienceComputer Science (R0)