Abstract
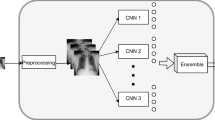
Since the beginning of the COVID-19 pandemic, more than 350 million cases and 5 million deaths have occurred. Since day one, multiple methods have been provided to diagnose patients who have been infected. Alongside the gold standard of laboratory analyses, deep learning algorithms on chest X-rays (CXR) have been developed to support the COVID-19 diagnosis. The literature reports that convolutional neural networks (CNNs) have obtained excellent results on image datasets when the tests are performed in cross-validation, but such models fail to generalize to unseen data. To overcome this limitation, we exploit the strength of multiple CNNs by building an ensemble of classifiers via an optimized late fusion approach. To demonstrate the system’s robustness, we present different experiments on open source CXR datasets to simulate a real-world scenario, where scans of patients affected by various lung pathologies and coming from external datasets are tested. Promising performances are obtained both in cross-validation and in external validation, obtaining an average accuracy of 93.02% and 91.02%, respectively.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Worldmeters coronavirus. https://www.worldometers.info/coronavirus/. Accessed 01 Feb 2022
Aljondi, R., et al.: Diagnostic value of imaging modalities for COVID-19: scoping review. J. Med. Internet Res. 22(8), e19673 (2020)
Basu, S., et al.: Deep learning for screening COVID-19 using chest x-ray images. In: 2020 IEEE Symposium Series on Computational Intelligence (SSCI), pp. 2521–2527. IEEE (2020)
Brown, G., et al.: Diversity creation methods: a survey and categorisation. Inf. Fusion 6(1), 5–20 (2005)
Brunese, L., et al.: Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from x-rays. Comput. Methods Programs Biomed. 196, 105608 (2020)
Cavalcanti, G.D., et al.: Combining diversity measures for ensemble pruning. Pattern Recogn. Lett. 74, 38–45 (2016)
Cipollari, S., et al.: Convolutional neural networks for automated classification of prostate multiparametric magnetic resonance imaging based on image quality. J. Magn. Reson. Imaging 55(2), 480–490 (2022)
Cohen, J.P., et al.: COVID-19 image data collection: prospective predictions are the future. arXiv preprint arXiv:2006.11988 (2020)
Das, D., Santosh, K.C., Pal, U.: Truncated inception net: COVID-19 outbreak screening using chest x-rays. Phys. Eng. Sci. Med. 43(3), 915–925 (2020). https://doi.org/10.1007/s13246-020-00888-x
Deng, J., et al.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. IEEE (2009)
Futoma, J., et al.: The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Health 2(9), e489–e492 (2020)
Guarrasi, V., et al.: A multi-expert system to detect COVID-19 cases in x-ray images. In: 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), pp. 395–400. IEEE (2021)
Guarrasi, V., et al.: Pareto optimization of deep networks for COVID-19 diagnosis from chest x-rays. Pattern Recogn. 121, 108242 (2022)
Hansen, L.K., et al.: Neural network ensembles. IEEE Trans. Pattern Anal. Mach. Intell. 12(10), 993–1001 (1990)
He, K., et al.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Huang, G., et al.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Iandola, F.N., et al.: SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and \(<\)0.5MB model size. arXiv preprint arXiv:1602.07360 (2016)
Jaeger, S., et al.: Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 4(6), 475 (2014)
Krizhevsky, A.: One weird trick for parallelizing convolutional neural networks. arXiv preprint arXiv:1404.5997 (2014)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Pereira, R.M., et al.: COVID-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 194, 105532 (2020)
Pouyanfar, S., et al.: A survey on deep learning: algorithms, techniques, and applications. ACM Comput. Surv. (CSUR) 51(5), 1–36 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sandler, M., et al.: MobileNetV2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018)
Shiraishi, J., et al.: Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 174(1), 71–74 (2000)
Simonyan, K., et al.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Soda, P., et al.: AIforCOVID: predicting the clinical outcomes in patients with COVID-19 applying AI to chest-x-rays. An Italian multicentre study. Med. Image Anal. 74, 102216 (2021)
Szegedy, C., et al.: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
Wang, L., et al.: Covid-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci. Rep. 10(1), 1–12 (2020)
Wang, X., et al.: ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2097–2106 (2017)
Wynants, L., et al.: Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. Brit. Med. J. 369 (2020)
Xie, S., et al.: Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1492–1500 (2017)
Zagoruyko, S., et al.: Wide residual networks. arXiv preprint arXiv:1605.07146 (2016)
Acknowledgements
This work is partially funded by: POR CAMPANIA FESR 2014–2020, AP1-OS1.3 project “Protocolli TC del torace a bassissima dose e tecniche di intelligenza artificiale per la diagnosi precoce e quantificazione della malattia da COVID-19” CUP D54I20001410002; EU project “University-Industrial Educational Centre in Advanced Biomedical and Medical Informatics (CeBMI) No. 612462-EPP-1-2019-1-SK-EPPKA2-KA”; “AI against COVID-19 Competition”, organized by IEEE SIGHT Montreal, Vision and Image Processing Research Group of the University of Waterloo, and DarwinAI Corp., and sponsored by Microsoft.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Guarrasi, V., Soda, P. (2022). Optimized Fusion of CNNs to Diagnose Pulmonary Diseases on Chest X-Rays. In: Sclaroff, S., Distante, C., Leo, M., Farinella, G.M., Tombari, F. (eds) Image Analysis and Processing – ICIAP 2022. ICIAP 2022. Lecture Notes in Computer Science, vol 13231. Springer, Cham. https://doi.org/10.1007/978-3-031-06427-2_17
Download citation
DOI: https://doi.org/10.1007/978-3-031-06427-2_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-06426-5
Online ISBN: 978-3-031-06427-2
eBook Packages: Computer ScienceComputer Science (R0)