Abstract
Glioblastoma is profoundly heterogeneous in regional microstructure and vasculature. Characterizing the spatial heterogeneity of glioblastoma could lead to more precise treatment. With unsupervised learning techniques, glioblastoma MRI-derived radiomic features have been widely utilized for tumor sub-region segmentation and survival prediction. However, the reliability of algorithm outcomes is often challenged by both ambiguous intermediate process and instability introduced by the randomness of clustering algorithms, especially for data from heterogeneous patients.
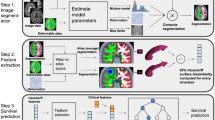
In this paper, we propose an adaptive unsupervised learning approach for efficient MRI intra-tumor partitioning and glioblastoma survival prediction. A novel and problem-specific Feature-enhanced Auto-Encoder (FAE) is developed to enhance the representation of pairwise clinical modalities and therefore improve clustering stability of unsupervised learning algorithms such as K-means. Moreover, the entire process is modelled by the Bayesian optimization (BO) technique with a custom loss function that the hyper-parameters can be adaptively optimized in a reasonably few steps. The results demonstrate that the proposed approach can produce robust and clinically relevant MRI sub-regions and statistically significant survival predictions.
C. Li—Equal contribution.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Behrens, T.E., et al.: Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn. Resonan. Med. Off. J. Int. Soc. Magn. Resonan. Med. 50(5), 1077–1088 (2003)
Beig, N., et al.: Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in glioblastoma. Clin. Cancer Res. 26(8), 1866–1876 (2020)
Brochu, E., Cora, V.M., De Freitas, N.: A tutorial on Bayesian optimization of expensive cost functions, with application to active user modeling and hierarchical reinforcement learning. arXiv preprint arXiv:1012.2599 (2010)
Dextraze, K., et al.: Spatial habitats from multiparametric MR imaging are associated with signaling pathway activities and survival in glioblastoma. Oncotarget 8(68), 112992 (2017)
Jenkinson, M., Bannister, P., Brady, M., Smith, S.: Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17(2), 825–841 (2002)
Leone, J., Zwenger, A.O., Leone, B.A., Vallejo, C.T., Leone, J.P.: Overall survival of men and women with breast cancer according to tumor subtype. Am. J. Clin. Oncol. 42(2), 215–220 (2019)
Li, C., et al.: Decoding the interdependence of multiparametric magnetic resonance imaging to reveal patient subgroups correlated with survivals. Neoplasia 21(5), 442–449 (2019)
Li, C., et al.: Multi-parametric and multi-regional histogram analysis of MRI: modality integration reveals imaging phenotypes of glioblastoma. Eur. Radiol. 29(9), 4718–4729 (2019)
Li, C., et al.: Intratumoral heterogeneity of glioblastoma infiltration revealed by joint histogram analysis of diffusion tensor imaging. Neurosurgery 85(4), 524–534 (2019)
Li, C., et al.: Low perfusion compartments in glioblastoma quantified by advanced magnetic resonance imaging and correlated with patient survival. Radiother. Oncol. 134, 17–24 (2019)
Mangla, R., et al.: Correlation between progression free survival and dynamic susceptibility contrast MRI perfusion in WHO grade III glioma subtypes. J. Neurooncol. 116(2), 325–331 (2013). https://doi.org/10.1007/s11060-013-1298-9
Meacham, C.E., Morrison, S.J.: Tumour heterogeneity and cancer cell plasticity. Nature 501(7467), 328–337 (2013)
Meilă, M.: Comparing clusterings by the variation of information. In: Schölkopf, B., Warmuth, M.K. (eds.) COLT-Kernel 2003. LNCS (LNAI), vol. 2777, pp. 173–187. Springer, Heidelberg (2003). https://doi.org/10.1007/978-3-540-45167-9_14
Meyer-Bäse, A., Saalbach, A., Lange, O., Wismüller, A.: Unsupervised clustering of fMRI and MRI time series. Biomed. Sig. Process. Control 2(4), 295–310 (2007)
Mohanty, A.K., Beberta, S., Lenka, S.K.: Classifying benign and malignant mass using GLCM and GLRLM based texture features from mammogram. Int. J. Eng. Res. Appl. 1(3), 687–693 (2011)
Nowosad, J., Stepinski, T.F.: Information theory as a consistent framework for quantification and classification of landscape patterns. Landscape Ecol. 34(9), 2091–2101 (2019). https://doi.org/10.1007/s10980-019-00830-x
Park, J.E., Kim, H.S., Kim, N., Park, S.Y., Kim, Y.H., Kim, J.H.: Spatiotemporal heterogeneity in multiparametric physiologic MRI is associated with patient outcomes in IDH-wildtype glioblastoma. Clin. Cancer Res. 27(1), 237–245 (2021)
Paszke, A., et al.: Pytorch: an imperative style, high-performance deep learning library. Adv. Neural. Inf. Process. Syst. 32, 8026–8037 (2019)
Patel, E., Kushwaha, D.S.: Clustering cloud workloads: K-means vs gaussian mixture model. Procedia Comput. Sci. 171, 158–167 (2020)
Pena, A., Green, H., Carpenter, T., Price, S., Pickard, J., Gillard, J.: Enhanced visualization and quantification of magnetic resonance diffusion tensor imaging using the p: q tensor decomposition. Br. J. Radiol. 79(938), 101–109 (2006)
Rasmussen, C.E.: Gaussian processes in machine learning. In: Bousquet, O., von Luxburg, U., Rätsch, G. (eds.) ML -2003. LNCS (LNAI), vol. 3176, pp. 63–71. Springer, Heidelberg (2004). https://doi.org/10.1007/978-3-540-28650-9_4
Sala, E., et al.: Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin. Radiol. 72(1), 3–10 (2017)
Smith, S.M., et al.: Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23, S208–S219 (2004)
Snoek, J., Larochelle, H., Adams, R.P.: Practical Bayesian optimization of machine learning algorithms. arXiv preprint arXiv:1206.2944 (2012)
Sottoriva, A., et al.: Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. 110(10), 4009–4014 (2013)
Syed, A.K., Whisenant, J.G., Barnes, S.L., Sorace, A.G., Yankeelov, T.E.: Multiparametric analysis of longitudinal quantitative MRI data to identify distinct tumor habitats in preclinical models of breast cancer. Cancers 12(6), 1682 (2020)
Van Griethuysen, J.J., et al.: Computational radiomics system to decode the radiographic phenotype. Can. Res. 77(21), e104–e107 (2017)
Von Luxburg, U.: Clustering stability: an overview. Found. Trends Mach. Learn. 2(3), 235–274 (2010)
Wu, J., et al.: Unsupervised clustering of quantitative image phenotypes reveals breast cancer subtypes with distinct prognoses and molecular pathways. Clin. Cancer Res. 23(13), 3334–3342 (2017)
Wu, J., Gong, G., Cui, Y., Li, R.: Intra-tumor partitioning and texture analysis of DCE-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J. Magn. Resonan. Imaging (JMRI) 44(5), 1107 (2016)
Xia, W., et al.: Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys. Med. Biol. 63(3), 035044 (2018)
Zhang, L.: Dirac delta function of matrix argument. Int. J. Theor. Phys. 1–28 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
5 Appendix
5 Appendix
1.1 5.1 Details of Dataset and Imagine Processing
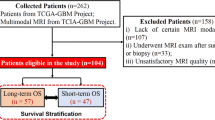
Patients with surgical resection (July 2010–August 2015) were consecutively recruited, with data prospectively collected by the multidisciplinary team (MDT) central review. All glioblastoma patients underwent pre-operative 3D MPRAGE (pre-contrast T1 and T1C), T2-weighted FLAIR, pMRI and dMRI sequences. All patients have a radiogical diagnosis of de novo glioblastoma, aged 18 to 75, eligible for craniotomy and radiotherapy, and all images resolution were resampled to \(1\times 1 \times 1\,\mathrm{{m}}^3\).
Co-registration of the images was accomplished using the linear registration tool (FLIRT) included in the Oxford Centre for Functional MRI of the Brain Software Library (FSL) v5.0.0 (Oxford, UK) [5, 23]. NordicICE was used to process dynamic susceptibility contrast (DSC), one of the most frequently utilised perfusion methods (NordicNeuroLab). The arterial input function was automatically defined. The diffusion toolbox in FSL was used to process the diffusion images (DTI) [1]. The isotropic (p) and anisotropic (q) components were computed after normalisation and eddy current correction [20].
1.2 Details for Clinical Features
In this study, through the BO, the tumor were divided into 5 sub-regions as \(\{\mathbf {P}_n\}_{n=1}^N\) from \(\{\mathbf {Z}_{m^{\prime }}\}_{m^{\prime }=1}^M\), the features processed by the well-trained FAE, where \(\mathbf {P}_n = \{\mathbf {p}_i\}_{i=1}^I\), \(\mathbf {p}_i \in \{1,2,3,4,5\}\) denotes the sub-region labels for each pixel. Rather than representing the numerical grey value of images, the value of each \(\mathbf {p}_i\) represents sub-region labels, rendering the majority of features in the GLCM and GLRLM families invalid. Finally, the Table 2 summarises the selected features which remain meaningful for the label matrix. Eventually, the clinical features \(\{\mathbf {F}_n \}_{n=1}^N\), where \(\mathbf {F}_n \in {\mathbb {R}}^{11 \times 1}\) include 9 spatial characteristics in Table 2 and the fraction of 2 significant sub-regions.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Li, Y., Li, C., Wei, Y., Price, S., Schönlieb, CB., Chen, X. (2022). Adaptive Unsupervised Learning with Enhanced Feature Representation for Intra-tumor Partitioning and Survival Prediction for Glioblastoma. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2021. Lecture Notes in Computer Science, vol 12962. Springer, Cham. https://doi.org/10.1007/978-3-031-08999-2_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-08999-2_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-08998-5
Online ISBN: 978-3-031-08999-2
eBook Packages: Computer ScienceComputer Science (R0)