Abstract
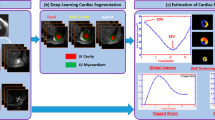
Myocardial strain is an important measure of cardiac performance, which can be altered when ejection fraction (EF) and other ventricular volumetric indices remain normal, providing an additional indicator for early detection of cardiac dysfunction. Cardiac tagging MRI is the gold standard for myocardial strain quantification but requires additional sequence acquisition and relatively complex post-processing procedures, which limit its clinical application. In this paper, we propose a framework for learning a joint latent representation of cine MRI and tagging MRI, such that tagging MRI can be synthesised and used to derive myocardial strain, given just cine MRI as inputs. Specifically, we use a multi-channel variational autoencoder to simultaneously learn features from tagging MRI and cine MRI, and project the information from these distinct channels into a common latent space to jointly analyse the multi-sequence data information. The inference process generates tagging MRI using only cine MRI as input, by conditionally sampling from the learned latent representation. Finally, automated tag tracking was performed using a cardiac motion tag tracking network on the generated tagging MRI, and myocardial strain was estimated. Experiments on the UK Biobank dataset show that our proposed framework can generate tagging images from cine images alone, and in turn, can be used to estimate myocardial strain effectively.
N. Ravikumar and A. F. Frangi—Joint last authors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ibrahim, E.-S.H.: Myocardial tagging by cardiovascular magnetic resonance: evolution of techniques–pulse sequences, analysis algorithms, and applications. J. Cardiovasc. Magn. Reson. 13(1), 1–40 (2011). https://doi.org/10.1186/1532-429X-13-36
Sutherland, G.R., Di Salvo, G., Claus, P., D'hooge, J., Bijnens, B.: Strain and strain rate imaging: a new clinical approach to quantifying regional myocardial function. J. Am. Soc. Echocardiogr. 17(7), 788–802 (2004). https://doi.org/10.1016/j.echo.2004.03.027
Wu, L., Germans, T., Güçlü, A., Heymans, M.W., Allaart, C.P., van Rossum, A.C.: Feature tracking compared with tissue tagging measurements of segmental strain by cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 16(1), 1–11 (2014). https://doi.org/10.1186/1532-429X-16-10
Götte, M.J., Germans, T., Rüssel, I.K., Zwanenburg, J.J., Marcus, J.T., van Rossum, A.C., et al.: Myocardial strain and torsion quantified by cardiovascular magnetic resonance tissue tagging: studies in normal and impaired left ventricular function 48(10), 2002–2011 (2006). https://doi.org/10.1016/j.jacc.2006.07.048
Guttman, M.A., Prince, J.L., McVeigh, E.R.: Tag and contour detection in tagged MR images of the left ventricle 13(1), 74–88 (1994). https://doi.org/10.1109/42.276146
Young, A.A., Kraitchman, D.L., Dougherty, L., Axel, L.: Tracking and finite element analysis of stripe deformation in magnetic resonance tagging. IEEE Trans. Med. Imaging 14(3), 413–421 (1995). https://doi.org/10.1109/42.414605
O’Shea, K., Nash, R.: An introduction to convolutional neural networks. arXiv preprint arXiv:1511.08458 (2015)
Pu, Y., Gan, Z., Henao, R., Yuan, X., Li, C., Stevens, A., et al.: Variational autoencoder for deep learning of images, labels and captions. In: 29th Conference on Neural Information Processing Systems (NIPS 2016), Barcelona, Spain, p. 29 (2016). https://doi.org/10.48550/arXiv.1609.08976
Antelmi, L., Ayache, N., Robert, P., Lorenzi, M.: Sparse multi-channel variational autoencoder for the joint analysis of heterogeneous data. In: International Conference on Machine Learning, Long Beach, California (2019)
Liu, X., Xing, F., Prince, J.L., Carass, A., Stone, M., El Fakhri, G., et al.: Dual-cycle constrained bijective VAE-GAN for tagged-to-cine magnetic resonance image synthesis. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), IEEE, France (2021). https://doi.org/10.1109/ISBI48211.2021.9433852
Loecher, M., Hannum, A.J., Perotti, L.E., Ennis, D.B.: Arbitrary point tracking with machine learning to measure cardiac strains in tagged MRI. In: Ennis, D.B., Perotti, L.E., Wang, V.Y. (eds.) FIMH 2021. LNCS, vol. 12738, pp. 213–222. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-78710-3_21
Biobank U: About uk biobank (2014)
Petersen, S.E., Matthews, P.M., Bamberg, F., Bluemke, D.A., Francis, J.M., Friedrich, M.G., et al.: Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank-rationale, challenges and approaches 15(1), 1–10 (2013). https://doi.org/10.1186/1532-429X-15-46
Zheng, Q., Delingette, H., Duchateau, N., Ayache, N.: 3-D consistent and robust segmentation of cardiac images by deep learning with spatial propagation. IEEE Trans. Med. Imaging 37(9), 2137–48 (2018). https://doi.org/10.48550/arXiv.1804.09400
Loecher, M., Perotti, L.E., Ennis, D.B.: Using synthetic data generation to train a cardiac motion tag tracking neural network. Med. Image Anal. 74,1022–1023 (2021). https://doi.org/10.1016/j.media.2021.102223
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., Fei-Fei, L.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami. IEEE (2009). https://doi.org/10.1109/CVPR.2009.5206848
Tran, D., Wang, H., Torresani, L., Ray, J., LeCun, Y., Paluri, M.: A closer look at spatiotemporal convolutions for action recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, USA, pp. 6450–6459 (2018)
Liu, R., Lehman, J., Molino, P., Petroski Such, F., Frank, E., Sergeev, A., et al.: An intriguing failing of convolutional neural networks and the coordconv solution, 31 (2018)
Loshchilov, I., Hutter, F.: SGDR: stochastic gradient descent with warm restarts. In: 5th International Conference on Learning Representations (ICLR 2016), Toulon, France (2016)
Prakash, K.B., Kanagachidambaresan, G.R. (eds.): Programming with TensorFlow. EICC, Springer, Cham (2021). https://doi.org/10.1007/978-3-030-57077-4
Wang, W., Lu, Y.: Analysis of the mean absolute error (MAE) and the root mean square error (RMSE) in assessing rounding model. In: IOP Conference Series: Materials Science and Engineering. IOP Publishing, (2018). https://doi.org/10.1088/1757-899X/324/1/012049
Dosselmann, R., Yang, X.D.: A comprehensive assessment of the structural similarity index. Signal Image Video Process. 5(1), 81–91 (2011). https://doi.org/10.1007/s11760-009-0144-1
Poobathy, D., Chezian, R.M.: Edge detection operators: peak signal to noise ratio based comparison. IJ Image Graph. Signal Process. 10, 55–61 (2014). https://doi.org/10.5815/ijigsp.2014.10.07
Bunce, C.: Correlation, agreement, and Bland–Altman analysis: statistical analysis of method comparison studies. Am. J. Ophthalmol. 148(1), 4–6 (2009). https://doi.org/10.1016/j.ajo.2008.09.032
Young, A.A., Li, B., Kirton, R.S., Cowan, B.R.: Generalized spatiotemporal myocardial strain analysis for DENSE and SPAMM imaging. Magn. Reson. Med. 67(6), 1590–159 (2012). https://doi.org/10.1002/mrm.23142
Acknowledgements
This research was conducted using data from the UK Biobank under access application 11350. AFF is funded by the Royal Academy of Engineering (INSILEX CiET1819\19), Engineering and Physical Sciences Research Council (EPSRC) programs TUSCA EP/V04799X/1, and the Royal Society Exchange Programme CROSSLINK IES\NSFC\201380, and was supported partly by China Scholarship Council Studentship with the University of Leeds.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Cheng, N., Bonazzola, R., Ravikumar, N., Frangi, A.F. (2022). A Generative Framework for Predicting Myocardial Strain from Cine-Cardiac Magnetic Resonance Imaging. In: Yang, G., Aviles-Rivero, A., Roberts, M., Schönlieb, CB. (eds) Medical Image Understanding and Analysis. MIUA 2022. Lecture Notes in Computer Science, vol 13413. Springer, Cham. https://doi.org/10.1007/978-3-031-12053-4_36
Download citation
DOI: https://doi.org/10.1007/978-3-031-12053-4_36
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-12052-7
Online ISBN: 978-3-031-12053-4
eBook Packages: Computer ScienceComputer Science (R0)