Abstract
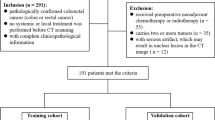
Although preoperative biopsy of rectal cancer (RC) is an essential step for confirmation of diagnosis, it currently fails to provide prognostic information to the clinician beyond a rough estimation of tumour grade. In this study we used a risk classification to stratified patient in low-risk and high-risk patients in relation to the disease free survival and the overall survival using histopathological post-operative features. The purpose of this study was to evaluate if low-risk and high-risk RC can be distinguished using a CT-based radiomics model. We retrospectively reviewed the preoperative abdominal contrast-enhanced CT of 40 patients with RC. CT portal-venous phase was used for manual RC segmentation by two radiologists. Radiomics parameters were extracted by using PyRadiomics (3.0) software, which automatically obtained a total of 120 radiomics features. An operator-independent statistical hybrid method was adopted for the selection and reduction of features, while discriminant analysis was used to construct the predictive model. Postoperative histopathological report was used as reference standard. Receiver operating characteristics (ROC) and areas under the ROC curve (AUROC) were calculated to evaluate the diagnostic performance of the most dis-criminating selected parameters. Sensitivity, specificity, and accuracy were calculated. In our study cohort, the original_shape_Maximum3DDiameter and origi-nal_shape_MajorAxisLength demonstrated a good performance in the differentiation between preoperative degree of RC, with an AUROC of 0.680%, sensitivity of 74.02%, specificity of 73.45%, positive predictive value of 81.47%, and accuracy of 73.71%. In conclusion, this preliminary analysis showed statistically significant differences in radiomics features between low-risk and high-risk RC.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Coppola, F., et al.: Radiomics and magnetic resonance imaging of rectal cancer: from engineering to clinical practice. Diagnostics (Basel, Switzerland) 11 (2021). https://doi.org/10.3390/DIAGNOSTICS11050756
Dekker, E., Tanis, P.J., Vleugels, J.L.A., Kasi, P.M., Wallace, M.B.: Colorectal cancer. Lancet 394, 1467–1480 (2019). https://doi.org/10.1016/S0140-6736(19)32319-0
Sauer, R., et al.: Preoperative versus postoperative chemoradiotherapy for rectal cancer. N. Engl. J. Med. 351, 1731–1740 (2004). https://doi.org/10.1056/NEJMoa040694
Paun, B.C., Cassie, S., MacLean, A.R., Dixon, E., Buie, W.D.: Postoperative complications following surgery for rectal cancer. Ann. Surg. 251, 807–818 (2010). https://doi.org/10.1097/SLA.0b013e3181dae4ed
Glynne-Jones, R., et al.: Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 28, iv22–iv40 (2017). https://doi.org/10.1093/annonc/mdx224
Balyasnikova, S., Brown, G.: Optimal imaging strategies for rectal cancer staging and ongoing management. Curr. Treat. Options Oncol. 17(6), 1–11 (2016). https://doi.org/10.1007/s11864-016-0403-7
Hou, M., Sun, J.H.: Emerging applications of radiomics in rectal cancer: state of the art and future perspectives. World J. Gastroenterol. 27, 3802–3814 (2021). https://doi.org/10.3748/wjg.v27.i25.3802
Park, K.-J., Choi, H.-J., Roh, M.-S., Kwon, H.-C., Kim, C.: Intensity of tumor budding and its prognostic implications in invasive colon carcinoma. Dis. Colon Rectum. 48, 1597–1602 (2005). https://doi.org/10.1007/s10350-005-0060-6
Vernuccio, F., Cannella, R., Comelli, A., Salvaggio, G., Lagalla, R., Midiri, M.: Radiomics and artificial intelligence: new frontiers in medicine. Recent. Prog. Med. 111(3), 130–135 (2020). Italian. https://doi.org/10.1701/3315.32853
Giambelluca, D., et al.: PI-RADS 3 lesions: role of prostate MRI texture analysis in the identification of prostate cancer. Curr. Probl. Diagn. Radiol. 50 (2019). https://doi.org/10.1067/J.CPRADIOL.2019.10.009
Cannella, R., la Grutta, L., Midiri, M., Bartolotta, T.V.: New advances in radiomics of gastrointestinal stromal tumors. World J. Gastroenterol. 26, 4729–4738 (2020). https://doi.org/10.3748/WJG.V26.I32.4729
Caruana, G., et al.: Texture analysis in susceptibility-weighted imaging may be useful to differentiate acute from chronic multiple sclerosis lesions. Eur. Radiol. 30(11), 6348–6356 (2020). https://doi.org/10.1007/s00330-020-06995-3
Cutaia, G., et al.: Radiomics and prostate MRI: current role and future applications. J. Imaging. 7, 34 (2021). https://doi.org/10.3390/jimaging7020034
Stefano, A., et al.: Performance of radiomics features in the quantification of idiopathic pulmonary fibrosis from HRCT. Diagnostics. 10, 306 (2020). https://doi.org/10.3390/diagnostics10050306
Stefano, A., et al.: Robustness of pet radiomics features: Impact of co-registration with MRI. Appl. Sci. 11, 10170 (2021). https://doi.org/10.3390/app112110170
Banna, G.L., et al.: Predictive and prognostic value of early disease progression by PET evaluation in advanced non-small cell lung cancer. Oncology 92, 39–47 (2016). https://doi.org/10.1159/000448005
Russo, G., et al.: Feasibility on the use of radiomics features of 11[C]-MET PET/CT in central nervous system tumours: preliminary results on potential grading discrimination using a machine learning model. Curr. Oncol. 28, 5318–5331 (2021). https://doi.org/10.3390/curroncol28060444
Li, M., et al.: A clinical-radiomics nomogram for the preoperative prediction of lymph node metastasis in colorectal cancer. J. Transl. Med. 18, 1 (2020). https://doi.org/10.1186/s12967-020-02215-0
Sun, Y., et al.: Radiomic features of pretreatment MRI could identify T stage in patients with rectal cancer: Preliminary findings. J. Magn. Reson. Imaging 48, 615–621 (2018). https://doi.org/10.1002/jmri.25969
Van Griethuysen, J.J.M., et al.: Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017). https://doi.org/10.1158/0008-5472.CAN-17-0339
Pyradiomics, https://pyradiomics.readthedocs.io/en/latest/features.html. Accessed 11 Apr 2022
Barone, S., et al.: Hybrid descriptive-inferential method for key feature selection in prostate cancer radiomics. Appl. Stoch. Model. Bus. Ind. 37, 961–972 (2021). https://doi.org/10.1002/asmb.2642
Alongi, P., et al.: Radiomics analysis of 18F-Choline PET/CT in the prediction of disease outcome in high-risk prostate cancer: an explorative study on machine learning feature classification in 94 patients. Eur. Radiol. 31(7), 4595–4605 (2021). https://doi.org/10.1007/s00330-020-07617-8
Salvaggio, G., et al.: Deep learning networks for automatic retroperitoneal sarcoma segmentation in computerized tomography. Appl. Sci. 12, 1665 (2022). https://doi.org/10.3390/app12031665
Stefano, A., Comelli, A.: Customized efficient neural network for covid-19 infected region identification in CT images. J. Imaging 7, 131 (2021). https://doi.org/10.3390/jimaging7080131
De Cecco, C.N., et al.: Texture analysis as imaging biomarker of tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3-T magnetic resonance. Invest. Radiol. 50, 239–245 (2015). https://doi.org/10.1097/RLI.0000000000000116
Liang, C., et al.: The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget 7, 31401–31412 (2016). https://doi.org/10.18632/ONCOTARGET.8919
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Canfora, I. et al. (2022). A Predictive System to Classify Preoperative Grading of Rectal Cancer Using Radiomics Features. In: Mazzeo, P.L., Frontoni, E., Sclaroff, S., Distante, C. (eds) Image Analysis and Processing. ICIAP 2022 Workshops. ICIAP 2022. Lecture Notes in Computer Science, vol 13373. Springer, Cham. https://doi.org/10.1007/978-3-031-13321-3_38
Download citation
DOI: https://doi.org/10.1007/978-3-031-13321-3_38
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13320-6
Online ISBN: 978-3-031-13321-3
eBook Packages: Computer ScienceComputer Science (R0)