Abstract
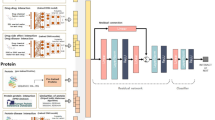
Predicting drug-target interactions plays an important role in shortening the cycle and reducing the cost of drug development. Although many existing approaches have been successful, most of them mainly start from one-dimensional sequences and do not take full advantage of the biological relationships between the drug and the target. In this paper, the heterogeneous networks are constructed based on biological properties such as drug, target, disease, side effects and their relationships. The features of drugs and targets are automatically learned using a neural network-based topology-preserving learning model. To improve the prediction accuracy of drug-target interactions, a RandSAS optimization strategy is designed, which introduces a stochastic factor based on drug-target binding affinity and drug-drug similarity to optimize the prediction process. The experimental result shows that the prediction results are relatively good when the similarity threshold is set to 0.6. In addition, based on the constructed heterogeneous network, RandSAS strategy could improve the accuracy of drug-target prediction to a certain extent.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dickson, M., Gagnon, J.P.: Key factors in the rising cost of new drug discovery and development. Nat. Rev. Drug Discov. 3, 417 (2004)
Pushpakom, S., et al.: Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41 (2018)
Tamimi, N.A., Ellis, P.: Drug development: from concept to marketing. Nephron Clin. Pract. 113, c125–c131 (2009)
Li, J., et al.: A survey of current trends in computational drug repositioning. Bioinformatics 17, 2–12 (2016)
Yang, M., Huang, L., Xu, Y., Lu, C., et al.: Heterogeneous graph inference with matrix completion for computational drug repositioning. Bioinformatics 36, 5456–5464 (2020)
Nosengo, N.: Can you teach old drugs new tricks? Nat. News 534, 314 (2016)
Cui, C., Ding, X., Wang, D., et al.: Drug repurposing against breast cancer by integrating drug-exposure expression profiles and drug–drug links based on graph neural network. Bioinformatics 37, 2930–2937 (2021)
Gottlieb, A., et al.: PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 7, 496 (2011)
Yang, L., Agarwal, P.: Systematic drug repositioning based on clinical side-effects. PLoS ONE 6, e28025 (2011)
Wang, Y., et al.: Drug repositioning by kernel-based integration of molecular structure, molecular activity, and phenotype data. PLoS ONE 8, e78518 (2013)
Oh, M., et al.: A network-based classification model for deriving novel drug-disease associations and assessing their molecular actions. PLoS ONE 9, e111668 (2014)
Wang, W., et al.: Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 30, 2923–2930 (2014)
Martínez, V., et al.: DrugNet: network-based drug-disease prioritization by integrating heterogeneous data. Artif. Intell. Med. 63, 41–49 (2015)
Luo, H., et al.: Drug repositioning based on comprehensive similarity measures and bi-random walk algorithm. Bioinformatics 32, 2664–2671 (2016)
Luo, H., et al.: Computational drug repositioning using low-rank matrix approximation and randomized algorithms. Bioinformatics 34, 1904–1912 (2018)
Zhang, W., et al.: Predicting drug-disease associations by using similarity constrained matrix factorization. Bioinformatics 19, 233 (2018)
Zhang, W., Xu, H., Li, X.Z., et al.: DRIMC: an improved drug repositioning approach using Bayesian inductive matrix completion. Bioinformatics 36(9), 2839–2847 (2020)
Lin, X.L., Zhang, X.L., Xu, X.: Efficient classification of hot spots and hub protein interfaces by recursive feature elimination and gradient boosting. IEEE/ACM Trans. Comput. Biol. Bioinf. 17(5), 1525–1534 (2020)
Lin, X.L., Zhang, X.L.: Identification of hot regions in hub protein-protein interactions by clustering and PPRA optimization. BMC Med. Inf. Decision Mak. 21(1), S1 (2021)
Wan, F., Hong, L., Xiao, A., et al.: NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug–target interactions. Bioinformatics 35, 104–111 (2019)
Luo, Y., Zhao, X., Zhou, J., et al.: A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 8, 573 (2017)
Knox, C., et al.: Drugbank 3.0: A comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 39, D1035–D1041 (2011)
Prasad, T.S.K., et al.: Human protein reference database-2009 update. Nucleic Acids Res. 37, D767–D772 (2009)
Davis, A.P., et al.: The comparative toxicogenomics database: update 2013. Nucleic Acids Res. 41, D1104–D1114 (2013)
Kuhn, M., Campillos, et al.: A side effect resource to capture phenotypic effects of drugs. Mol. Sydt. Biol. 6, 343 (2009)
Acknowledgements
The authors thank the members of Machine Learning and Artificial Intelligence Laboratory, School of Computer Science and Technology, Wuhan University of Science and Technology, for their helpful discussion within seminars. This work was supported by the National Innovation and Entrepreneurship Training Program for University Students (202110488019) and the National Natural Science Foundation of China (No. 61972299).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Xiang, H., Li, A., Lin, X. (2022). An Optimization Method for Drug-Target Interaction Prediction Based on RandSAS Strategy. In: Huang, DS., Jo, KH., Jing, J., Premaratne, P., Bevilacqua, V., Hussain, A. (eds) Intelligent Computing Theories and Application. ICIC 2022. Lecture Notes in Computer Science, vol 13394. Springer, Cham. https://doi.org/10.1007/978-3-031-13829-4_47
Download citation
DOI: https://doi.org/10.1007/978-3-031-13829-4_47
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13828-7
Online ISBN: 978-3-031-13829-4
eBook Packages: Computer ScienceComputer Science (R0)