Abstract
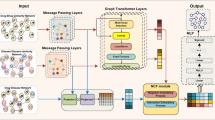
Exploring the association between drugs and diseases can help to accelerate the process of drug development to a certain extent. In order to investigate the association between drugs and diseases, this paper constructs a network composed of different types of nodes, and proposes a model NSAP based on neighborhood subgraph prediction. The model captures local and global information around the target node through metagraphs and contextual graphs, respectively, and can generate node representations with rich information. In addition, in metagraphs and context diagrams, the model takes advantage of graph structures to automatically generate weights for edges, which better reflects the degree of association of different neighbor nodes with the target node. At last, the attention mechanism is used to aggregate the nodal representations generated by different metapaths in the graph, so that the final representation of the nodes incorporates different semantic information. For the edge prediction, a correlation score between drug-disease node pairs is calculated by the decoder. The experimental results have confirmed that our model does have certain effect by comparing it with state of the art method. The data and code are available at: https://github.com/jqq125/NSAP.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Additional Files
All additional files are available at: https://github.com/jqq125/NSAP
References
Ashburn, T.T., Thor, K.B.: Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3(8), 673–683 (2004)
Jarada, T.N., Rokne, J.G., Alhajj, R.: A review of computational drug repositioning: strategies, approaches, opportunities, challenges, and directions. J. Cheminform. 12(1), 1–23 (2020). https://doi.org/10.1186/s13321-020-00450-7
Li, J., et al.: A survey of current trends in computational drug repositioning. Brief. Bioinform. 17(1), 2–12 (2015). https://doi.org/10.1093/bib/bbv020
Sadeghi, S.S., Keyvanpour, M.R.: An analytical review of computational drug repurposing. IEEE/ACM Trans. Comput. Biol. Bioinform. 1–1 (2019)
Perozzi, B., et al.: DeepWalk: online learning of social representations. In: Macskassy, S.A. et al. (eds.) The 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD 2014, 24–27 August 2014. pp. 701–710. ACM, New York, NY, USA (2014). https://doi.org/10.1145/2623330.2623732
Grover, A., Leskovec, J.: Node2vec: scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 855–864. Association for Computing Machinery, New York, NY, USA (2016). https://doi.org/10.1145/2939672.2939754
Liu, H. et al.: Inferring new indications for approved drugs via random walk on drug-disease heterogenous networks. BMC Bioinformatics. 17, 17, 539 (2016). https://doi.org/10.1186/s12859-016-1336-7
Yang, C., et al.: Network representation learning with rich text information. In: Proceedings of the 24th International Conference on Artificial Intelligence, pp. 2111–2117. AAAI Press (2015)
Cao, S., et al.: GraRep: learning graph representations with global structural information. In: Proceedings of the 24th ACM International on Conference on Information and Knowledge Management, pp. 891–900 Association for Computing Machinery, New York, NY, USA (2015). https://doi.org/10.1145/2806416.2806512
Liu, Y., et al.: Neighborhood regularized logistic matrix factorization for drug-target interaction prediction. PLOS Comput. Biol. 12 (2016)
N. Kipf, T., Welling, M.: Semi-Supervised Classification with Graph Convolutional Networks. ICLR (2017). https://doi.org/10.48550/arXiv.1609.02907
Velickovic, P., et al.: Graph attention networks. In: 6th International Conference on Learning Representations, ICLR 2018, Vancouver, BC, Canada, April 3–May 3 2018, Conference Track Proceedings. OpenReview.net (2018)
Hamilton, W.L., et al.: Inductive representation learning on large graphs. In: NIPS. (2017)
Wishart, D.S., et al.: DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, Database-Issue, D1074–D1082 (2018). https://doi.org/10.1093/nar/gkx1037
Bateman, A., et al.: UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49(Database-Issue), D480–D489 (2021). https://doi.org/10.1093/nar/gkaa1100
Rappaport, N., et al.: MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 45(Database-Issue), D877–D887 (2017). https://doi.org/10.1093/nar/gkw1012
Brown, A.S., Patel, C.J.: A standard database for drug repositioning. Sci. Data. 4, 170029 (2017)
González, J.P., et al.: The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48(Database-Issue) D845–D855 (2020). https://doi.org/10.1093/nar/gkz1021
Huser, V., et al.: ClinicalTrials.gov: Adding Value through Informatics. In: AMIA 2015, American Medical Informatics Association Annual Symposium, 14–18 Nov 2015. AMIA, San Francisco, CA, USA (2015)
Dong, Y., et al.: metapath2vec: scalable representation learning for heterogeneous networks. In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 13–17 August 2017, pp. 135–144. ACM, Halifax, NS, Canada (2017). https://doi.org/10.1145/3097983.3098036
Wang, X., et al.: Heterogeneous Graph Attention Network. WWW 2019, The Web Conference 2019, 13–17 May 2019, pp. 2022–2032. ACM, San Francisco, CA, USA, (2019). https://doi.org/10.1145/3308558.3313562
Fu, X., et al.: MAGNN: Metapath Aggregated Graph Neural Network for Heterogeneous Graph Embedding. WWW 2020: The Web Conference 2020, 20–24 April 2020, pp. 2331–2341. ACM/IW3C2Taipei, Taiwan (2020). https://doi.org/10.1145/3366423.3380297
He, M., et al.: Factor graph-aggregated heterogeneous network embedding for disease-gene association prediction. BMC Bioinf. 22(1), 165 (2021). https://doi.org/10.1186/s12859-021-04099-3
Acknowledgements
This work was supported by the grants from the National Key R&D Program of China (2021YFA0910700), Shenzhen science and technology university stable support program (GXWD20201230155427003-20200821222112001), Shenzhen Science and Technology Program (JCYJ20200109113201726), Guangdong Basic and Applied Basic Research Foundation (2021A1515012461 and 2021A1515220115).
Author information
Authors and Affiliations
Contributions
QJ designed the study, performed bioinformatics analysis and drafted the manuscript. All of the authors performed the analysis and participated in the revision of the manuscript. JL and YW conceived of the study, participated in its design and coordination and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
The authors declare that they have no competing interests.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jiao, Q., Jiang, Y., Zhang, Y., Wang, Y., Li, J. (2022). NSAP: A Neighborhood Subgraph Aggregation Method for Drug-Disease Association Prediction. In: Huang, DS., Jo, KH., Jing, J., Premaratne, P., Bevilacqua, V., Hussain, A. (eds) Intelligent Computing Theories and Application. ICIC 2022. Lecture Notes in Computer Science, vol 13394. Springer, Cham. https://doi.org/10.1007/978-3-031-13829-4_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-13829-4_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13828-7
Online ISBN: 978-3-031-13829-4
eBook Packages: Computer ScienceComputer Science (R0)