Abstract
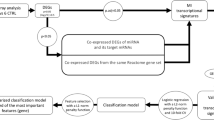
Acute myocardial infarction (AMI) is a severe disease that can occur in all age groups. About 8.5 million patients die of this disease every year. Although the diagnostic technology of AMI is relatively mature, there are still many limitations. We aim to use comprehensive bioinformatics and machine learning algorithms to study the potential molecular mechanism of acute myocardial infarction and seek new prevention and treatment strategies. Methods: The expression profiles of GSE66360 and GSE48060 were downloaded from the Gene Expression Omnibus database, microarray datasets were integrated, and differential genes were obtained to be further analyzed by bioinformatic technique. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, Disease Oncology (DO) analysis and Gene Set Enrichment Analysis (GSEA) were performed on the differential genes using R software, respectively. Then the Lasso algorithm was used to identify the AMI-related essential genes in the training set and validate them in the test set. Potential mechanistic analyses of the development of AMI included the following: the expression differences of crucial genes, differences in immune cell infiltration, immune cell correlation, and the correlation between critical genes and immune cells between normal and AMI samples. Results: Finally, five essential genes were screened, including CLEC4D, CSF3R, SLC11A1, CLEC12A, and TAGAP. The expression of critical genes differed between normal, and AMI samples and the genes can be used as a diagnostic factor in patients. Meanwhile, normal and AMI samples showed significant differences in immune infiltration, and the expression of critical genes was closely related to the abundance of immune cell infiltration. Conclusion: In this study, five essential genes were screened, and the underlying molecular mechanisms of AMI pathogenesis were analyzed, which may provide theoretical support for the diagnosis, prevention, prognosis evaluation and targeted immune therapy of AMI patients.
Z. Zhan and T. Zhao—Contributed to the work equally
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Thygesen, K., Alpert, J.S., Jaffe, A.S.: Third universal definition of myocardial infarction. Eur. Heart J. 50, 2173–2195 (2012)
Libby, P.: Mechanisms of acute coronary syndromes and their implications for therapy. N. Engl. J. Med. 368, 2004–2013 (2013)
White, H.D., Chew, D.P.: Acute myocardial infarction. Lancet. Cardiol. Clin. 2, 79–94 (1984)
Yeh, R.W., Sidney, S., Chandra, M., Sorel, M., Selby, J.V., Go, A.S.: Population trends in the incidence and outcomes of acute myocardial infarction. New England J. Med. 362, 2155–2165 (2010)
Murray, C.J.L., et al.: Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990–2013: Quantifying the epidemiological transition (2015)
Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the global burden of disease study 2010. Lancet (London, England) 380, 2197 (2012)
French, J.K., Hellkamp, A.S., Armstrong, P.W., Eric, C.: Mechanical complications after percutaneous coronary intervention in ST-elevation myocardial infarction (from APEX-AMI). American J. Cardiol. 105 (2010)
Cai, W., Li, H., Zhang, Y., Han, G.: Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. PeerJ 8, e8390 (2020)
Zhang, X., Zhang, W., Jiang, Y., Liu, K., Song, F.: Identification of functional lncRNAs in gastric cancer by integrative analysis of GEO and TCGA data. J. Cellular Biochem. 120 (2019)
Suzuki, T., Kano, S., Suzuki, M., Yasukawa, S., Homma, A.: Enhanced angiogenesis in salivary duct carcinoma ex-pleomorphic adenoma. Front. Oncol. 10, 603717 (2021)
Tibshirani, R.: Regression shrinkage and selection via the lasso. J. Royal Statist. Soc. Ser. B (Methodological) 58 (1996)
Barrett, T., et al.: NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 39, 1005–1010 (2013)
Meltzer, D.P.S.: GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 (2007)
Ritchie, M.E., et al.: Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015)
Yu, G., Wang, L.G., Han, Y., He, Q.Y.: clusterProfiler: an r package for comparing biological themes among gene clusters. Omics-a J. Integ. Biol. 16, 284–287 (2012)
Robin, X., et al.: pROC: an open-source package for r and s+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011)
Deng, Y.J., Ren, E.H., Yuan, W.H., Zhang, G.Z., Xie, Q.Q.: GRB10 and E2F3 as diagnostic markers of osteoarthritis and their correlation with immune infiltration. Diagnostics 10, 171 (2020)
Weintraub, W.S., et al.: Value of primordial and primary prevention for cardiovascular disease: a policy statement from the american heart association. Circulation 124, 967 (2011)
Bruyninckx, R., Aertgeerts, B., Bruyninckx, P., Buntinx, F.: Signs and symptoms in diagnosing acute myocardial infarction and acute coronary syndrome: a diagnostic meta-analysis. Br. J. Gen. Pract. 58, 105–111 (2008)
Asari, P., et al.: Acute myocardial infarction hospital admissions and deaths in England: A national follow-back and follow-forward record-linkage study. Lancet Public Health 2, e191 (2017)
Xu, J.Y., Xiong, Y.Y., Lu, X.T., Yang, Y.J.: Regulation of type 2 immunity in myocardial infarction. Other 10 (2019)
Weil, B.R., Neelamegham, S.: Selectins and immune cells in acute myocardial infarction and post-infarction ventricular remodeling: pathophysiology and novel treatments. Front. Immunol. 10 (2019)
Suresh, R., et al.: Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. J. Mol. Cell. Cardiol. 74, 13–21 (2014)
Sun, J., et al.: Deficiency of antigen-presenting cell invariant chain reduces atherosclerosis in mice. Circulation 122, 808–820 (2010)
Tobin, S.W., Alibhai, F.J., Weisel, R.D., Li, R.K.: Considering cause and effect of immune cell aging on cardiac repair after myocardial infarction. Cells. 9 (1894)
Suykens, J., Vandewalle, J.: Least squares support vector machine classifiers. Neural Process. Lett. 9, 293–300 (1999)
Singh, N., Gedda, M.R., Tiwari, N., Singh, S.P., Bajpai, S., Singh, R.K.: Solute carrier protein family 11 member 1 (Slc11a1) activation efficiently inhibits leishmania donovani survival in host macrophages. J. Parasitic Diseases 41 (2017)
Franco, M., et al.: Slc11a1 (Nramp1) alleles interact with acute inflammation loci to modulate wound-healing traits in mice. Mammalian Genome Off. J. Int. Mammalian Genome Soc. 18, 263 (2007)
Marcelo, D.F., et al.: Pristane-induced arthritis loci interact with the Slc11a1 gene to determine susceptibility in mice selected for high inflammation. PLoS ONE 9, e88302 (2014)
Friedman, M.A., Choi, D., Planck, S.R., Rosenbaum, J.T., Sibley, C.H.: Gene expression pathways across multiple tissues in antineutrophil cytoplasmic antibody-associated vasculitis reveal core pathways of disease pathology. J. Rheumatol. 46 (2019)
Lechermeier, C.G., D’Orazio, A., Romanos, M., Lillesaar, C., Drepper, C.: Distribution of transcripts of the GFOD gene family members gfod1 and gfod2 in the zebrafish central nervous system. Gene Expr. Patterns 36, 119111 (2020)
Franzke, A., et al.: G-CSF as immune regulator in t cells expressing the g-CSF receptor: Implications for transplantation and autoimmune diseases. Blood 102, 734–739 (2003)
Morris, K.T., et al.: G-CSF and g-CSFR are highly expressed in human gastric and colon cancers and promote carcinoma cell proliferation and migration. Br. J. Cancer 110, 1211–1220 (2014)
Maria, A., Attya, B., Peter, J., Yang, Z.: Identification and in silico analysis of functional SNPs of human TAGAP protein: a comprehensive study. PLoS ONE 13, e0188143 (2018)
Graham, L.M., et al.: The c-type lectin receptor CLECSF8 (CLEC4D) is expressed by myeloid cells and triggers cellular activation through SYK kinase. J. Biol. Chem. 287, 25964–25974 (2012)
Newby, A.C.: Pathogenesis of atherosclerosis. Principles; Practice of Geriatric Medicine (1949)
Carbone, F., Nencioni, A., Mach, F., Vuilleumier, N., Montecucco, F.: Pathophysiological role of neutrophils in acute myocardial infarction. Thrombosis and Haemostasis (2017)
Nahrendorf, M., Pittet, M.J., Swirski, F.K.: Basic science for clinicians monocytes: protagonists of infarct inflammation and repair after myocardial infarction (2019)
Nikolaos, G., Frangogiannis: regulation of the inflammatory response in cardiac repair. Circulation research (2012)
Carbone, F., Nencioni, A., Mach, F., Vuilleumier, N., Montecucco, F.: Pathophysiological role of neutrophils in acute myocardial infarction. Thromb. Haemost. 109, 501–514 (2013)
Nahrendorf, M., Swirski, F.K.: Regulating repair: regulatory t cells in myocardial infarction. Circ. Res. 115, 7–9 (2014)
Acknowledgement
This study was supported by Provincial Science and Technology Grant of Shanxi Province (20210302124588), Science and technology innovation project of Shanxi province universities (2019L0683).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Zhan, Z., Zhao, T., Bi, X., Yang, J., Han, P. (2022). Identification and Evaluation of Key Biomarkers of Acute Myocardial Infarction by Machine Learning. In: Huang, DS., Jo, KH., Jing, J., Premaratne, P., Bevilacqua, V., Hussain, A. (eds) Intelligent Computing Theories and Application. ICIC 2022. Lecture Notes in Computer Science, vol 13394. Springer, Cham. https://doi.org/10.1007/978-3-031-13829-4_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-13829-4_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13828-7
Online ISBN: 978-3-031-13829-4
eBook Packages: Computer ScienceComputer Science (R0)