Abstract
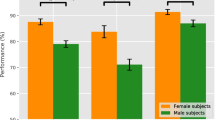
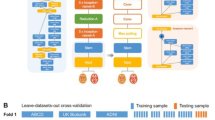
Convolutional neural networks have enabled significant improvements in medical image-based diagnosis. It is, however, increasingly clear that these models are susceptible to performance degradation when facing spurious correlations and dataset shift, leading, e.g., to underperformance on underrepresented patient groups. In this paper, we compare two classification schemes on the ADNI MRI dataset: a simple logistic regression model using manually selected volumetric features, and a convolutional neural network trained on 3D MRI data. We assess the robustness of the trained models in the face of varying dataset splits, training set sex composition, and stage of disease. In contrast to earlier work in other imaging modalities, we do not observe a clear pattern of improved model performance for the majority group in the training dataset. Instead, while logistic regression is fully robust to dataset composition, we find that CNN performance is generally improved for both male and female subjects when including more female subjects in the training dataset. We hypothesize that this might be due to inherent differences in the pathology of the two sexes. Moreover, in our analysis, the logistic regression model outperforms the 3D CNN, emphasizing the utility of manual feature specification based on prior knowledge, and the need for more robust automatic feature selection.
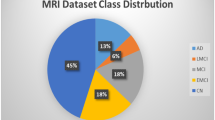
Data used in preparation of this article was obtained from the Alzheimers Disease Neuroimaging Initiative (ADNI) database (http://www.adni-info.org/). The investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but did not participate in analysis or writing of this report.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abrol, A., et al.: Deep learning encodes robust discriminative neuroimaging representations to outperform standard machine learning. Nat. Commun. 12(1), 1–7 (2021). https://doi.org/10.1038/s41467-020-20655-6
Adragna, R., Creager, E., Madras, D., Zemel, R.: Fairness and robustness in invariant learning: A case study in toxicity classification. In: NeurIPS Workshop on Algorithmic Fairness through the Lens of Causality and Interpretability (2020). https://arxiv.org/abs/2011.06485
Arjovsky, M., Bottou, L., Gulrajani, I., Lopez-Paz, D.: Invariant risk minimization. arXiv (2019). https://arxiv.org/abs/1907.02893
Ashburner, J.: SPM: a history. Neuroimage 62(2), 791–800 (2012). https://doi.org/10.1016/j.neuroimage.2011.10.025
Azulay, A., Weiss, Y.: Why do deep convolutional networks generalize so poorly to small image transformations? J. Mach. Learn. Res. 20(184), 1–25 (2019). http://jmlr.org/papers/v20/19-519.html
Banerjee, I., et al.: Reading race: AI recognises patient’s racial identity in medical images. arXiv (2021). https://arxiv.org/abs/2107.10356
Cowling, T.E., Cromwell, D.A., Bellot, A., Sharples, L.D., van der Meulen, J.: Logistic regression and machine learning predicted patient mortality from large sets of diagnosis codes comparably. J. Clin. Epidemiol. 133, 43–52 (2021). https://doi.org/10.1016/j.jclinepi.2020.12.018
D’Amour, A., et al.: Underspecification presents challenges for credibility in modern machine learning. CoRR (2020). https://arxiv.org/abs/2011.03395
Falcon, W.: The PyTorch Lightning team: PyTorch Lightning (version 1.5.9) (2019). https://www.pytorchlightning.ai
Fernández, A., García, S., Galar, M., Prati, R.C., Krawczyk, B., Herrera, F.: Performance measures. In: Learning from Imbalanced Data Sets, pp. 47–61. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-98074-4_3
Fischl, B.: Freesurfer. Neuroimage 62(2), 774–781 (2012). https://doi.org/10.1016/j.neuroimage.2012.01.021
Geirhos, R., et al.: Shortcut learning in deep neural networks. Nat. Mach. Intell. 2(11), 665–673 (2020). https://doi.org/10.1038/s42256-020-00257-z
Jack, C.R., et al.: The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Resonan. Imaging 27(4), 685–691 (2008). https://doi.org/10.1002/jmri.21049
Jacobucci, R., Littlefield, A.K., Millner, A.J., Kleiman, E.M., Steinley, D.: Evidence of inflated prediction performance: a commentary on machine learning and suicide research. Clin. Psychol. Sci. 9(1), 129–134 (2021). https://doi.org/10.1177/2167702620954216
Larrazabal, A.J., Nieto, N., Peterson, V., Milone, D.H., Ferrante, E.: Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl. Acad. Sci. 117(23), 12592–12594 (2020). https://doi.org/10.1073/pnas.1919012117
Malone, I.B., et al.: Accurate automatic estimation of total intracranial volume: a nuisance variable with less nuisance. NeuroImage 104, 366–372 (2015). https://doi.org/10.1016/j.neuroimage.2014.09.034
Mielke, M., Vemuri, P., Rocca, W.: Clinical epidemiology of Alzheimer’s disease: assessing sex and gender differences. Clin. Epidemiol. 6, 37 (2014). https://doi.org/10.2147/clep.s37929
Nusinovici, S., et al.: Logistic regression was as good as machine learning for predicting major chronic diseases. J. Clin. Epidemiol. 122, 56–69 (2020). https://doi.org/10.1016/j.jclinepi.2020.03.002
Obermeyer, Z., Powers, B., Vogeli, C., Mullainathan, S.: Dissecting racial bias in an algorithm used to manage the health of populations. Science 366(6464), 447–453 (2019). https://doi.org/10.1126/science.aax2342
Pawlowski, N., Castro, D.C., Glocker, B.: Deep structural causal models for tractable counterfactual inference. In: Advances in Neural Information Processing Systems, vol. 33, pp. 857–869. Curran Associates, Inc. (2020), https://proceedings.neurips.cc/paper/2020/file/0987b8b338d6c90bbedd8631bc499221-Paper.pdf
Pérez-García, F., Sparks, R., Ourselin, S.: TorchIO: a python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Comput. Methods Programs Biomed. 208, 106236 (2021). https://doi.org/10.1016/j.cmpb.2021.106236
Podcasy, J.L., Epperson, C.N.: Considering sex and gender in Alzheimer disease and other dementias. Dialogues Clin. Neurosc. 18(4), 437–446 (2016). https://doi.org/10.31887/dcns.2016.18.4/cepperson
Quiñonero-Candela, J., Sugiyama, M., Lawrence, N.D., Schwaighofer, A.: Dataset Shift in Machine Learning. MIT Press, Cambridge (2009)
Seyyed-Kalantari, L., Zhang, H., McDermott, M.B.A., Chen, I.Y., Ghassemi, M.: Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat. Med. 27(12), 2176–2182 (2021). https://doi.org/10.1038/s41591-021-01595-0
Tinauer, C., et al.: Interpretable brain disease classification and relevance-guided deep learning. medRxiv (2021). https://doi.org/10.1101/2021.09.09.21263013
Varoquaux, G., et al.: Assessing and tuning brain decoders: cross-validation, caveats, and guidelines. NeuroImage 145, 166–179 (2017). https://doi.org/10.1016/j.neuroimage.2016.10.038
Wen, J., et al.: Convolutional neural networks for classification of Alzheimer’s disease: overview and reproducible evaluation. Med. Image Anal. 63, 101694 (2020). https://doi.org/10.1016/j.media.2020.101694
Wynants, L., et al.: Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal. BMJ 369, m1328 (2020). https://doi.org/10.1136/bmj.m1328
Yi, P.H., et al.: Radiology “forensics”: determination of age and sex from chest radiographs using deep learning. Emerg. Radiol. 28(5), 949–954 (2021). https://doi.org/10.1007/s10140-021-01953-y
Zhao, Q., Adeli, E., Pohl, K.M.: Training confounder-free deep learning models for medical applications. Nat. Commun. 11(1), 1–9 (2020). https://doi.org/10.1038/s41467-020-19784-9
Acknowledgements
We thank Morten Rieger Hannemose for helpful comments on the manuscript and the statistical analysis. This research was supported by Danmarks Frie Forskningsfond (9131-00097B), the Novo Nordisk Foundation through the Center for Basic Machine Learning Research in Life Science (NNF20OC0062606) and the Pioneer Centre for AI, DNRF grant number P1. Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from private sector institutions. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles.
Author information
Authors and Affiliations
Consortia
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Petersen, E. et al. (2022). Feature Robustness and Sex Differences in Medical Imaging: A Case Study in MRI-Based Alzheimer’s Disease Detection. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13431. Springer, Cham. https://doi.org/10.1007/978-3-031-16431-6_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-16431-6_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16430-9
Online ISBN: 978-3-031-16431-6
eBook Packages: Computer ScienceComputer Science (R0)