Abstract
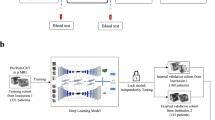
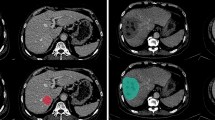
Wavelets have shown significant promise for medical image decomposition and artifact pre-processing by representing inputs via shifted and scaled components of a specified mother wavelet function. However, wavelets could also be leveraged within deep neural networks as activation functions for neurons (called wavelons) in the hidden layer. Integrating wavelons into a convolutional neural network architecture (termed a “wavelon network” (WN)) offers additional flexibility and stability during optimization, but the resulting model complexity has caused it to be limited to low-dimensional applications. Towards addressing these issues, we present the Residual Wavelon Convolutional Network (RWCN), a novel integrated WN architecture that employs weighted skip connections (to enable residual learning) together with image convolutions and wavelet activation functions to more efficiently capture high-dimensional disease response-specific patterns from medical imaging data. In addition to developing the analytical basis for wavelet activation functions as used in this work, we implemented RWCNs by adapting the popular VGG and ResNet architectures. Evaluation was conducted within three different challenging clinical problems: (a) predicting pathologic complete response (pCR) to neoadjuvant chemoradiation via 153 pre-treatment T2-weighted (T2w) MRI scans in rectal cancers, (b) evaluating pCR after chemoradiation via 100 post-treatment T2w MRIs in rectal cancers, as well as (c) risk stratifying patients who will or will not require surgery after aggressive medication in Crohn’s disease using 73 baseline MRI scans. In comparison to 4 state-of-the-art alternative models (VGG-16, VGG-19, ResNet-18, ResNet-50), RWCN architectures yielded significantly improved and more efficient classifier performance on unseen data in multi-institutional validation cohorts (hold-out accuracies of 0.82, 0.85, and 0.88, respectively).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Billings, S.A.: Nonlinear System Identification: NARMAX Methods in the Time, Frequency, and Spatio-Temporal Domains. Wiley, Hoboken (2013)
Gu, J., Yang, T.S., Ye, J.C., Yang, D.H.: CycleGAN denoising of extreme low-dose cardiac CT using wavelet-assisted noise disentanglement. Med. Image Anal. 74, 102209 (2021)
Chen, L., et al.: Super-resolved enhancing and edge deghosting (SEED) for spatiotemporally encoded single-shot MRI. Med. Image Anal. 23(1), 1–14 (2015)
Lao, J., et al.: A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci. Rep. 7(1), 1–8 (2017)
Liu, J., Li, P., Tang, X., Li, J., Chen, J.: Research on improved convolutional wavelet neural network. Sci. Rep. 11(1), 1–14 (2021)
Zaeemzadeh, A., Rahnavard, N., Shah, M.: Norm-preservation: why residual networks can become extremely deep? IEEE Trans. Pattern Anal. Mach. Intell. 43(11), 3980–3990 (2020)
Li, Q., Shen, L., Guo, S., Lai, Z.: Wavecnet: wavelet integrated CNNs to suppress aliasing effect for noise-robust image classification. IEEE Trans. Image Process. 30, 7074–7089 (2021)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Xie, H., et al.: Cross-attention multi-branch network for fundus diseases classification using SLO images. Med. Image Anal. 71, 102031 (2021)
Bruna, J., Mallat, S.: Invariant scattering convolution networks. IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1872–1886 (2013)
Wiatowski, T., Bölcskei, H.: A mathematical theory of deep convolutional neural networks for feature extraction. IEEE Trans. Inf. Theor. 64(3), 1845–1866 (2017)
Rodriguez, M.X.B., et al.: Deep adaptive wavelet network. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 3111–3119 (2020)
Gu, J., et al.: Recent advances in convolutional neural networks. Pattern Recogn. 77, 354–377 (2018)
Alexandridis, A.K., Zapranis, A.D.: Wavelet Neural Networks: with Applications in Financial Engineering, Chaos, and Classification. Wiley, Hoboken (2014)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Biswas, K., Kumar, S., Banerjee, S., Pandey, A.K.: Tanhsoft-dynamic trainable activation functions for faster learning and better performance. IEEE Access 9, 120613–120623 (2021)
Naitzat, G., Zhitnikov, A., Lim, L.H.: Topology of deep neural networks. J. Mach. Learn. Res. 21(184), 1–40 (2020)
Oyedotun, O.K., Al Ismaeil, K., Aouada, D.: Why is everyone training very deep neural network with skip connections? IEEE Trans. Neural Netw. Learn. Syst. (2022)
Furusho, Y., Ikeda, K.: Theoretical analysis of skip connections and batch normalization from generalization and optimization perspectives. APSIPA Trans. Sig. Inf. Process. 9 (2020)
Lundberg, S.M., et al.: From local explanations to global understanding with explainable AI for trees. Nature Mach. Intell. 2(1), 56–67 (2020)
Lundberg, S.M., et al.: Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nature Biomed. Eng. 2(10), 749 (2018)
Ancona, M., Oztireli, C., Gross, M.: Explaining deep neural networks with a polynomial time algorithm for Shapley value approximation. In: International Conference on Machine Learning, pp. 272–281. PMLR (2019)
Acknowledgments
Research supported by NCI (1U01CA248226-01), DOD/CDMRP (W81XWH-21-1-0345), and NIDDK (1F31DK130587-01A1). Content solely responsibility of the authors and does not necessarily represent the official views of the NIH, DOD, or the United States Government.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Sadri, A.R., DeSilvio, T., Chirra, P., Singh, S., Viswanath, S.E. (2022). Residual Wavelon Convolutional Networks for Characterization of Disease Response on MRI. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13433. Springer, Cham. https://doi.org/10.1007/978-3-031-16437-8_35
Download citation
DOI: https://doi.org/10.1007/978-3-031-16437-8_35
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16436-1
Online ISBN: 978-3-031-16437-8
eBook Packages: Computer ScienceComputer Science (R0)