Abstract
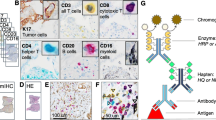
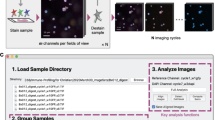
Multispectral immunofluorescence (M-IF) analysis is used to investigate the cellular landscape of tissue sections and spatial interaction of cells. However, complex makeup of markers in the images hinders the accurate quantification of cell phenotypes. We developed DeepMIF, a new deep learning (DL) based tool with a graphical user interface (GUI) to detect and quantify cell phenotypes on M-IF images, and visualize whole slide image (WSI) and cell phenotypes. To identify cell phenotypes, we detected cells on the deconvoluted images followed by co-expression analysis to classify cells expressing single or multiple markers. We trained, tested and validated our model on \(>50k\) expert single-cell annotations from multiple immune panels on 15 samples of follicular lymphoma patients. Our algorithm obtained a cell classification accuracy and area under the curve (AUC) \(\ge 0.98\) on an independent validation panel. The cell phenotype identification took on average 27.5 min per WSI, and rendering of the WSI took on average 0.07 minutes. DeepMIF is optimized to run on local computers or high-performance clusters independent of the host platform. These suggest that the DeepMIF is an accurate and efficient tool for the analysis and visualization of M-IF images, leading to the identification of novel prognostic cell phenotypes in tumours.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Tan, W.C.C., et al.: Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 40(4), 135–153 (2020)
Bortolomeazzi, M., et al.: A simpli (single-cell identification from multiplexed images) approach for spatially resolved tissue phenotypingat single-cell resolution. bioRxiv (2021)
Yu, W., et al.: A preliminary study of deep-learning algorithm for analyzing multiplex immunofluorescence biomarkers in body fluid cytology specimens. Acta Cytol. 65(4), 348–353 (2021)
Hoyt, C.C.: Multiplex immunofluorescence and multispectral imaging: forming the basis of a clinical test platform for immuno-oncology. Front. Mol. Biosci. 8, 442 (2021)
Lin, J.-R.: Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 7, e31657 (2018)
Pulsawatdi, A.V., et al.: A robust multiplex immunofluorescence and digital pathology workflow for the characterisation of the tumour immune microenvironment. Mol. Oncol. 14(10), 2384–2402 (2020)
Hagos, Y.B., et al.: High inter-follicular spatial co-localization of CD8+ FOXP3+ with CD4+ CD8+ cells predicts favorable outcome in follicular lymphoma. Hematol. Oncol. (2022)
Dimitriou, N., Arandjelović, O., Caie, P.D.: Deep learning for whole slide image analysis: an overview. Front. Med. 6, 264 (2019)
Hagos, Y.B., Narayanan, P.L., Akarca, A.U., Marafioti, T., Yuan, Y.: ConCORDe-net: cell count regularized convolutional neural network for cell detection in multiplex immunohistochemistry images. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11764, pp. 667–675. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32239-7_74
Sanchez, K., et al.: Multiplex immunofluorescence to measure dynamic changes in tumor-infiltrating lymphocytes and pd-l1 in early-stage breast cancer. Breast Can. Res. 23(1), 1–15 (2021)
Lee, C.-W., Ren, Y.J., Marella, M., Wang, M., Hartke, J., Couto, S.S.: Multiplex immunofluorescence staining and image analysis assay for diffuse large b cell lymphoma. J. Immunol. Methods 478, 112714 (2020)
Maric, D., et al.: Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks. Nat. Commun. 12(1), 1–12 (2021)
Ghahremani, P., et al.: Deep learning-inferred multiplex immunofluorescence for ihc image quantification. bioRxiv, Deepliif (2021)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Shu, M.: Deep learning for image classification on very small datasets using transfer learning (2019)
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, pp. 249–256. JMLR Workshop and Conference Proceedings (2010)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 92014)
Acknowledgment
Y.H.B received funding from European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie (No766030). Y.Y acknowledges funding from Cancer Research UK Career Establishment Award (C45982/A21808), Breast Cancer Now (2015NovPR638), Children’s Cancer and Leukaemia Group (CCLGA201906), NIH U54 CA217376 and R01 CA185138, CDMRP Breast Cancer Research Program Award BC132057, CRUK Brain Tumour Awards (TARGET-GBM), European Commission ITN (H2020-MSCA-ITN-2019), Wellcome Trust (105104/Z/14/Z), and The Royal Marsden/ICR National Institute of Health Research Biomedical Research Centre. GG and RLR are supported by Gilead Fellowship Program (Ed. 2017). T.M is supported by the UK National Institute of Health Research University College London Hospital Biomedical Research Centre. A.U.A is supported by Cancer Research UK-UCL Centre Cancer Immuno-therapy Accelerator Award.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Hagos, Y.B. et al. (2022). DeepMIF: Deep Learning Based Cell Profiling for Multispectral Immunofluorescence Images with Graphical User Interface. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13434. Springer, Cham. https://doi.org/10.1007/978-3-031-16440-8_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-16440-8_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16439-2
Online ISBN: 978-3-031-16440-8
eBook Packages: Computer ScienceComputer Science (R0)