Abstract
This work proposes a self-supervised algorithm to segment each arbitrary anatomical structure in a 3D medical image produced under various acquisition conditions, dealing with domain shift problems and generalizability. Furthermore, we advocate an interactive setting in the inference time, where the self-supervised model trained on unlabeled volumes should be directly applicable to segment each test volume given the user-provided single slice annotation. To this end, we learn a novel 3D registration network, namely Vol2Flow, from the perspective of image sequence registration to find 2D displacement fields between all adjacent slices within a 3D medical volume together. Specifically, we present a novel 3D CNN-based architecture that finds a series of registration flows between consecutive slices within a whole volume, resulting in a dense displacement field. A new self-supervised algorithm is proposed to learn the transformations or registration fields between the series of 2D images of a 3D volume. Consequently, we enable gradually propagating the user-provided single slice annotation to other slices of a volume in the inference time. We demonstrate that our model substantially outperforms related methods on various medical image segmentation tasks through several experiments on different medical image segmentation datasets. Code is available at https://github.com/AdelehBitarafan/Vol2Flow.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
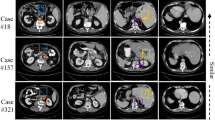
Similar content being viewed by others
References
Ahmad, M., et al.: Deep belief network modeling for automatic liver segmentation. IEEE Access 7, 20585–20595 (2019)
Arganda-Carreras, I., et al.: Non-rigid consistent registration of 2D image sequences. Phys. Med. Biol. 55(20), 6215 (2010)
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38(8), 1788–1800 (2019)
Bitarafan, A., Baghshah, M.S., Gheisari, M.: Incremental evolving domain adaptation. IEEE Trans. Knowl. Data Eng. 28(8), 2128–2141 (2016)
Bitarafan, A., Nikdan, M., Baghshah, M.S.: 3D image segmentation with sparse annotation by self-training and internal registration. IEEE J. Biomed. Health Inform. 25(7), 2665–2672 (2020)
Canny, J.: A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 6, 679–698 (1986)
Chen, S., Bortsova, G., García-Uceda Juárez, A., van Tulder, G., de Bruijne, M.: Multi-task attention-based semi-supervised learning for medical image segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 457–465. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32248-9_51
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Conze, P.H., et al.: Abdominal multi-organ segmentation with cascaded convolutional and adversarial deep networks. Artif. Intell. Med. 117, 102109 (2021)
Farnebäck, G.: Two-frame motion estimation based on polynomial expansion. In: Bigun, J., Gustavsson, T. (eds.) SCIA 2003. LNCS, vol. 2749, pp. 363–370. Springer, Heidelberg (2003). https://doi.org/10.1007/3-540-45103-X_50
Heller, N., et al.: Data from c4kc-kits [data set]. Cancer Imaging Arch. 10 (2019)
Hermann, S., Werner, R.: High accuracy optical flow for 3D medical image registration using the census cost function. In: Klette, R., Rivera, M., Satoh, S. (eds.) PSIVT 2013. LNCS, vol. 8333, pp. 23–35. Springer, Heidelberg (2014). https://doi.org/10.1007/978-3-642-53842-1_3
Hesamian, M.H., Jia, W., He, X., Kennedy, P.: Deep learning techniques for medical image segmentation: achievements and challenges. J. Digit. Imaging 32(4), 582–596 (2019)
Kavur, A.E., et al.: Chaos challenge-combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 69, 101950 (2021)
Keeling, S.L., Ring, W.: Medical image registration and interpolation by optical flow with maximal rigidity. J. Math. Imaging Vis. 23(1), 47–65 (2005)
Li, Z., Dong, Z., Yu, A., He, Z., Zhu, X.: A robust image sequence registration algorithm for videosar combining surf with inter-frame processing. In: IGARSS 2019–2019 IEEE International Geoscience and Remote Sensing Symposium, pp. 2794–2797. IEEE (2019)
Liu, X., Song, L., Liu, S., Zhang, Y.: A review of deep-learning-based medical image segmentation methods. Sustainability 13(3), 1224 (2021)
Mocanu, S., Moody, A.R., Khademi, A.: FlowREG: fast deformable unsupervised medical image registration using optical flow. arXiv preprint arXiv:2101.09639 (2021)
Radiuk, P.: Applying 3D U-net architecture to the task of multi-organ segmentation in computed tomography. Appl. Comput. Syst. 25(1), 43–50 (2020)
Roth, H., Farag, A., Turkbey, E., Lu, L., Liu, J., Summers, R.: Data from pancreas-CT (2016)
Roth, H., et al.: A new 2.5 d representation for lymph node detection in CT. Cancer Imaging Arch. (2018)
Soler, L., et al.: 3D image reconstruction for comparison of algorithm database: a patient specific anatomical and medical image database. Technical report, IRCAD, Strasbourg, France (2010)
Van Ginneken, B., Heimann, T., Styner, M.: 3D segmentation in the clinic: a grand challenge. In: MICCAI workshop on 3D segmentation in the clinic: a grand challenge, vol. 1, pp. 7–15 (2007)
Wang, G., et al.: Interactive medical image segmentation using deep learning with image-specific fine tuning. IEEE Trans. Med. Imaging 37(7), 1562–1573 (2018)
Wang, G., et al.: Slic-Seg: slice-by-slice segmentation propagation of the placenta in fetal MRI using one-plane scribbles and online learning. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 29–37. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_4
Xia, Y., et al.: 3D semi-supervised learning with uncertainty-aware multi-view co-training. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 3646–3655 (2020)
Yeung, P.-H., Namburete, A.I.L., Xie, W.: Sli2Vol: annotate a 3D volume from a single slice with self-supervised learning. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12902, pp. 69–79. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87196-3_7
Zhang, X., Xie, W., Huang, C., Zhang, Y., Wang, Y.: Self-supervised tumor segmentation through layer decomposition. arXiv preprint arXiv:2109.03230 (2021)
Zheng, Z., Zhang, X., Xu, H., Liang, W., Zheng, S., Shi, Y.: A unified level set framework combining hybrid algorithms for liver and liver tumor segmentation in CT images. In: BioMed research International 2018 (2018)
Acknowledgements
The authors were partially supported by the grant NPRP-11S-1219- 170106 from the Qatar National Research Fund (a member of the Qatar Foundation). The findings herein are however solely the responsibility of the authors.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Bitarafan, A., Azampour, M.F., Bakhtari, K., Soleymani Baghshah, M., Keicher, M., Navab, N. (2022). Vol2Flow: Segment 3D Volumes Using a Sequence of Registration Flows. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13434. Springer, Cham. https://doi.org/10.1007/978-3-031-16440-8_58
Download citation
DOI: https://doi.org/10.1007/978-3-031-16440-8_58
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16439-2
Online ISBN: 978-3-031-16440-8
eBook Packages: Computer ScienceComputer Science (R0)