Abstract
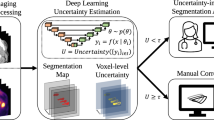
Bayesian Neural Nets (BNN) are increasingly used for robust organ auto-contouring. Uncertainty heatmaps extracted from BNNs have been shown to correspond to inaccurate regions. To help speed up the mandatory quality assessment (QA) of contours in radiotherapy, these heatmaps could be used as stimuli to direct visual attention of clinicians to potential inaccuracies. In practice, this is non-trivial to achieve since many accurate regions also exhibit uncertainty. To influence the output uncertainty of a BNN, we propose a modified accuracy-versus-uncertainty (AvU) metric as an additional objective during model training that penalizes both accurate regions exhibiting uncertainty as well as inaccurate regions exhibiting certainty. For evaluation, we use an uncertainty-ROC curve that can help differentiate between Bayesian models by comparing the probability of uncertainty in inaccurate versus accurate regions. We train and evaluate a FlipOut BNN model on the MICCAI2015 Head and Neck Segmentation challenge dataset and on the DeepMind-TCIA dataset, and observed an increase in the AUC of uncertainty-ROC curves by 5.6% and 5.9%, respectively, when using the AvU objective. The AvU objective primarily reduced false positives regions (uncertain and accurate), drawing less visual attention to these regions, thereby potentially improving the speed of error detection.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Alex Kendall, V.B., Cipolla, R.: Bayesian SegNet: model uncertainty in deep convolutional encoder-decoder architectures for scene understanding. In: Proceedings of the British Machine Vision Conference (BMVC). BMVA Press (2017)
Arega, T.W., Bricq, S., Meriaudeau, F.: Leveraging uncertainty estimates to improve segmentation performance in cardiac MR. In: Sudre, C.H., et al. (eds.) UNSURE/PIPPI -2021. LNCS, vol. 12959, pp. 24–33. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87735-4_3
Blundell, C., Cornebise, J., Kavukcuoglu, K., Wierstra, D.: Weight uncertainty in neural network. In: International Conference on Machine Learning. PMLR (2015)
Bragman, F.J.S., et al.: Uncertainty in multitask learning: joint representations for probabilistic MR-only radiotherapy planning. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_1
Brouwer, C.L., et al.: 3D variation in delineation of head and neck organs at risk. Radiat. Oncol. 7(1), 1–10 (2012)
Camarasa, R., et al.: Quantitative comparison of Monte-Carlo dropout uncertainty measures for multi-class segmentation. In: Sudre, C.H., et al. (eds.) UNSURE/GRAIL -2020. LNCS, vol. 12443, pp. 32–41. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-60365-6_4
Chen, Z., et al.: A novel hybrid convolutional neural network for accurate organ segmentation in 3D head and neck CT images. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 569–578. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_54
Gal, Y.: Uncertainty in deep learning. Ph.D. thesis, University of Cambridge (2016)
Gal, Y., Ghahramani, Z.: Dropout as a Bayesian approximation: representing model uncertainty in deep learning. In: International Conference on Machine Learning, pp. 1050–1059. PMLR (2016)
Guo, C., Pleiss, G., Sun, Y., Weinberger, K.Q.: On calibration of modern neural networks. In: International Conference on Machine Learning, pp. 1321–1330. PMLR (2017)
Islam, M., Glocker, B.: Spatially varying label smoothing: capturing uncertainty from expert annotations. In: Feragen, A., Sommer, S., Schnabel, J., Nielsen, M. (eds.) IPMI 2021. LNCS, vol. 12729, pp. 677–688. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-78191-0_52
Iwamoto, S., Raytchev, B., Tamaki, T., Kaneda, K.: Improving the reliability of semantic segmentation of medical images by uncertainty modeling with Bayesian deep networks and curriculum learning. In: Sudre, C.H., et al. (eds.) UNSURE/PIPPI -2021. LNCS, vol. 12959, pp. 34–43. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87735-4_4
Krishnan, R., Tickoo, O.: Improving model calibration with accuracy versus uncertainty optimization. In: Advances in Neural Information Processing Systems (2020)
Laves, M.H., Ihler, S., Kortmann, K.P., Ortmaier, T.: Well-calibrated model uncertainty with temperature scaling for dropout variational inference. arXiv preprint arXiv:1909.13550 (2019)
Mehrtash, A., Wells, W.M., Tempany, C.M., Abolmaesumi, P., Kapur, T.: Confidence calibration and predictive uncertainty estimation for deep medical image segmentation. IEEE Trans. Med. Imaging 39, 3868–3878 (2020)
Mobiny, A., Yuan, P., Moulik, S.K., Garg, N., Wu, C.C., Van Nguyen, H.: DropConnect is effective in modeling uncertainty of Bayesian deep networks. Sci. Rep. 11(1), 1–14 (2021)
Mody, P.P., de Plaza, N.C., Hildebrandt, K., van Egmond, R., de Ridder, H., Staring, M.: Comparing Bayesian models for organ contouring in head and neck radiotherapy. In: Medical Imaging 2022: Image Processing. International Society for Optics and Photonics, SPIE (2022)
Monteiro, M., et al.: Stochastic segmentation networks: modelling spatially correlated aleatoric uncertainty. Adv. Neural Inf. Process. Syst. 33, 12756–12767 (2020)
Mukhoti, J., Gal, Y.: Evaluating Bayesian deep learning methods for semantic segmentation. CoRR arXiv:abs/1811.12709 (2018)
Nair, T., Precup, D., Arnold, D.L., Arbel, T.: Exploring uncertainty measures in deep networks for multiple sclerosis lesion detection and segmentation. Med. Image Anal. 59, 101557 (2020)
Nikolov, S., et al.: Clinically applicable segmentation of head and neck anatomy for radiotherapy: deep learning algorithm development and validation study. J. Med. Internet Res. 23(7), e26151 (2021)
Raudaschl, P.F., et al.: Evaluation of segmentation methods on head and neck CT: auto-segmentation challenge 2015. Med. Phys. 44(5), 2020–2036 (2017)
Sander, J., de Vos, B.D., Išgum, I.: Automatic segmentation with detection of local segmentation failures in cardiac MRI. Sci. Rep. 10, 21769 (2020)
Sander, J., de Vos, B.D., Wolterink, J.M., Išgum, I.: Towards increased trustworthiness of deep learning segmentation methods on cardiac MRI. In: Medical Imaging 2019: Image Processing, vol. 10949, pp. 324–330. SPIE (2019)
Soberanis-Mukul, R.D., Navab, N., Albarqouni, S.: Uncertainty-based graph convolutional networks for organ segmentation refinement. In: Medical Imaging with Deep Learning, pp. 755–769. PMLR (2020)
Van Dijk, L.V., et al.: Improving automatic delineation for head and neck organs at risk by deep learning contouring. Radiother. Oncol. 142, 115–123 (2020)
van der Veen, J., Gulyban, A., Nuyts, S.: Interobserver variability in delineation of target volumes in head and neck cancer. Radiotherapy Oncol. 137, 9–15 (2019)
Wen, Y., Vicol, P., Ba, J., Tran, D., Grosse, R.: Flipout: efficient pseudo- independent weight perturbations on mini-batches. In: Proceedings of the 6th International Conference on Learning Representations (2018)
Acknowledgements
The research for this work was funded by Varian, a Siemens Healthineers Company, through the HollandPTC-Varian Consortium (grant id 2019022) and partly financed by the Surcharge for Top Consortia for Knowledge and Innovation (TKIs) from the Ministry of Economic Affairs and Climate, The Netherlands.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Mody, P., Chaves-de-Plaza, N.F., Hildebrandt, K., Staring, M. (2022). Improving Error Detection in Deep Learning Based Radiotherapy Autocontouring Using Bayesian Uncertainty. In: Sudre, C.H., et al. Uncertainty for Safe Utilization of Machine Learning in Medical Imaging. UNSURE 2022. Lecture Notes in Computer Science, vol 13563. Springer, Cham. https://doi.org/10.1007/978-3-031-16749-2_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-16749-2_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16748-5
Online ISBN: 978-3-031-16749-2
eBook Packages: Computer ScienceComputer Science (R0)