Abstract
Batch normalization (BN) uniformly shifts and scales the activations based on the statistics of a batch of images. However, the intensity distribution of the background pixels often dominates the BN statistics because the background accounts for a large proportion of the entire image. This paper focuses on enhancing BN with the intensity distribution of foreground pixels, the one that really matters for image segmentation. We propose a new normalization strategy, named categorical normalization (CateNorm), to normalize the activations according to categorical statistics. The categorical statistics are obtained by dynamically modulating specific regions in an image that belong to the foreground. CateNorm demonstrates both precise and robust segmentation results across five public datasets obtained from different domains, covering complex and variable data distributions. It is attributable to the ability of CateNorm to capture domain-invariant information from multiple domains (institutions) of medical data.
Code is available at https://github.com/lambert-x/CateNorm.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ba, J.L., Kiros, J.R., Hinton, G.E.: Layer normalization (2016)
Bloch, N., et al.: NCI-ISBI 2013 challenge: automated segmentation of prostate structures. The Cancer Imaging Archive (2015). http://doi.org/10.7937/K9/TCIA.2015.zF0vlOPv
Chang, W.G., You, T., Seo, S., Kwak, S., Han, B.: Domain-specific batch normalization for unsupervised domain adaptation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 7354–7362 (2019)
Chen, J., et al.: TransUNet: transformers make strong encoders for medical image segmentation (2021)
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11211, pp. 833–851. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01234-2_49
Clark, K., et al.: The cancer imaging archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26(6), 1045–1057 (2013)
Dumoulin, V., Shlens, J., Kudlur, M.: A learned representation for artistic style. In: 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, 24–26 April 2017, Conference Track Proceedings. OpenReview.net (2017). https://openreview.net/forum?id=BJO-BuT1g
Fu, S., et al.: Domain adaptive relational reasoning for 3D multi-organ segmentation. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12261, pp. 656–666. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59710-8_64
Gibson, E., et al.: Multi-organ abdominal CT reference standard segmentations, February 2018. https://doi.org/10.5281/zenodo.1169361
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Hu, S., Yuan, J., Wang, S.: Cross-modality synthesis from MRI to pet using adversarial U-Net with different normalization. In: 2019 International Conference on Medical Imaging Physics and Engineering (ICMIPE), pp. 1–5. IEEE (2019)
Huang, X., Belongie, S.: Arbitrary style transfer in real-time with adaptive instance normalization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1501–1510 (2017)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: International Conference on Machine Learning, pp. 448–456. PMLR (2015)
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: No new-net. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 234–244. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_21
Kao, P.-Y., Ngo, T., Zhang, A., Chen, J.W., Manjunath, B.S.: Brain tumor segmentation and tractographic feature extraction from structural MR images for overall survival prediction. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 128–141. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_12
Landman, B., Xu, Z., Igelsias, J., Styner, M., Langerak, T., Klein, A.: 2015 MICCAI multi-atlas labeling beyond the cranial vault workshop and challenge (2015). https://doi.org/10.7303/syn3193805
Lemaître, G., Martí, R., Freixenet, J., Vilanova, J.C., Walker, P.M., Meriaudeau, F.: Computer-aided detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: a review. Comput. Biol. Med. 60, 8–31 (2015)
Li, Y., Wang, N., Shi, J., Liu, J., Hou, X.: Revisiting batch normalization for practical domain adaptation (2016)
Liu, Q., Dou, Q., Yu, L., Heng, P.A.: MS-Net: multi-site network for improving prostate segmentation with heterogeneous MRI data. IEEE Trans. Med. Imaging 39(9), 2713–2724 (2020)
Park, T., Liu, M.Y., Wang, T.C., Zhu, J.Y.: Semantic image synthesis with spatially-adaptive normalization. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2337–2346 (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Roth, H., Farag, A., Turkbey, E.B., Lu, L., Liu, J., Summers, R.M.: Data from Pancreas-CT (2016). https://doi.org/10.7937/K9/TCIA.2016.TNB1KQBU, The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.tNB1kqBU
Roth, H.R., et al.: DeepOrgan: multi-level deep convolutional networks for automated pancreas segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 556–564. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24553-9_68
Ulyanov, D., Vedaldi, A., Lempitsky, V.: Instance normalization: the missing ingredient for fast stylization (2017)
Wu, Y., He, K.: Group normalization. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11217, pp. 3–19. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01261-8_1
Xia, X., Kulis, B.: W-Net: a deep model for fully unsupervised image segmentation (2017)
Yu, L., Yang, X., Chen, H., Qin, J., Heng, P.A.: Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MR images. In: Thirty-First AAAI Conference on Artificial Intelligence (2017)
Zhou, X.Y., Yang, G.Z.: Normalization in training U-Net for 2-D biomedical semantic segmentation. IEEE Robot. Autom. Lett. 4(2), 1792–1799 (2019)
Acknowledgments
This work was supported by the Lustgarten Foundation for Pancreatic Cancer Research. We also thank Quande Liu for the discussion.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendices
A Details of Aligning Input Distribution Algorithm
Assume that we have N source domains \(S_1, S_2, S_3, ..., S_N\), with \(M_1, M_2, M_3, ..., M_N\) examples respectively, where the i-th domain source domain \(S_i\) consists of an image set \(\{\textbf{x}_{i,j}\in \mathbb {R}^{D_{i,j}}\}_{j=1,...,M_i}\) as well as their associated annotations. Our goal is to align the image distributions of these source domains with the target domain T based on the class-wise (region-wise) statistics. The algorithm can be illustrated as the following steps:
Step 1: Calculate class-wise statistics of each case
Firstly, we calculate the mean and standard deviation of each case in both the source domain and the target domain.
where \(\textbf{x}_{i,j}^{c}\) denotes the pixels which belong to the c-th class (region) in image \(\textbf{x}_{i,j}\), with the number of pixels denoted as \(|D^c_{i,j}|\). As a special case, \(i=T\) indicates the target domain.
Step 2: Estimate aligned (new) class-wise statistics
Next, we calculate the mean of the statistics over all examples obtained in each domain as follows:
Based on the \(\bar{\mu }_{i}^{c}\), we now estimate the new class-wise mean \(\tilde{\mu }_{i,j}\) for each case of the source domain \(S_i\) as follows:
where \(M_T\) denotes the number of cases in the target domain T. Similarly, the new standard deviation \(\tilde{\sigma }_{i,j}\) can be computed by:
Step 3: Align each case with the estimated statistics
Based on the computed new mean and standard deviation \(\tilde{\mu }_{i,j}\), \(\tilde{\sigma }_{i,j}\), the aligned image \(\tilde{\textbf{x}}_{i,j}\) can be computed as:
B Implementation Details
C Details of the prostate datasets
(See Tables 6).
D Training procedure of CateNorm

E Average Surface Distance (ASD) Comparison
The detailed average surface distance results of both prostate segmentation and abdominal segmentation tasks can be found in Tables 7 and 8. The proposed CateNorm achieves the lowest average ASD on both tasks, even under the more challenging multi-domain setting (Tables 9, 10, 12).
Performance gain under partial annotation. We compare our method to the baseline with fewer annotated classes (i.e., 3/5). We can see that by partitioning the images into different number of regions, CateNorm consistently achieves better results than BN for all tested organs. This suggests that our algorithm is not sensitive to the number of regions.
Qualitative results comparison. We compare our baseline and the other SOTA method under the multi-domain setting on prostate segmentation and abdominal multi-organ segmentation. Results in the first three rows clearly show that our method outperforms others as their results are cracked and incomplete with these unapparent prostate boundaries. And the results in the last two rows show our methods could better suppress inconsistent class information inside a close segmented area (e.g., reducing false positives inside the stomach) and predict hard organs like the pancreas more accurately by incorporating general and categorical statistics.
Set CateNorm block(s) early. This table compares performance with single CateNorm block set in different positions. Adding the CateNorm to the encoder (block index 1–5) always yields better performance than adding to the decoder (block index 6–10). In general, the performance decreases as the block index increases. We believe that it is because the earlier layers in the encoder extract lower-level features that are less discriminative than the decoder features.
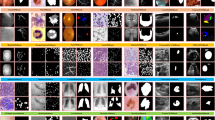
CateNorm does normalize with semantic information. This figure visualizes the learned \(\gamma ^{CateNorm }\) (1st row) and \(\beta ^{CateNorm }\) (2nd row) of a CateNorm layer in a CateNorm block on different channels of the intermediate CateNorm layer during the second forward. With prior class information as guidance, CateNorm can modulate spatially-adaptive parameters. Such spatial-wise modulation can be complementary to the channel-wise modulation accomplished by BN, and derives more discriminative features that benefit segmentation.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Xiao, J. et al. (2022). CateNorm: Categorical Normalization for Robust Medical Image Segmentation. In: Kamnitsas, K., et al. Domain Adaptation and Representation Transfer. DART 2022. Lecture Notes in Computer Science, vol 13542. Springer, Cham. https://doi.org/10.1007/978-3-031-16852-9_13
Download citation
DOI: https://doi.org/10.1007/978-3-031-16852-9_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16851-2
Online ISBN: 978-3-031-16852-9
eBook Packages: Computer ScienceComputer Science (R0)