Abstract
Human anatomy, morphology, and associated diseases can be studied using medical imaging data. However, access to medical imaging data is restricted by governance and privacy concerns, data ownership, and the cost of acquisition, thus limiting our ability to understand the human body. A possible solution to this issue is the creation of a model able to learn and then generate synthetic images of the human body conditioned on specific characteristics of relevance (e.g., age, sex, and disease status). Deep generative models, in the form of neural networks, have been recently used to create synthetic 2D images of natural scenes. Still, the ability to produce high-resolution 3D volumetric imaging data with correct anatomical morphology has been hampered by data scarcity and algorithmic and computational limitations. This work proposes a generative model that can be scaled to produce anatomically correct, high-resolution, and realistic images of the human brain, with the necessary quality to allow further downstream analyses. The ability to generate a potentially unlimited amount of data not only enables large-scale studies of human anatomy and pathology without jeopardizing patient privacy, but also significantly advances research in the field of anomaly detection, modality synthesis, learning under limited data, and fair and ethical AI. Code and trained models are available at: https://github.com/AmigoLab/SynthAnatomy.
G. Novati and M. Vella—Work done while at NVIDIA.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Implementation used: https://github.com/lucidrains/performer-pytorch.
References
Sudlow, C., et al.: UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12(3) (2015)
Clifford, R., Jack Jr., et al.: The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27(4), 685–691 (2008)
Simpson, A.L., et al.: A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 (2019)
Chong, C.K., Ho, E.T.W.: Synthesis of 3D MRI brain images with shape and texture generative adversarial deep neural networks. IEEE Access 9, 64747–64760 (2021)
Lin, W., et al.: Bidirectional mapping of brain MRI and pet with 3D reversible GAN for the diagnosis of Alzheimer’s disease. Front. Neurosci. 15, 357 (2021)
Rusak, F., et al.: 3D Brain MRI GAN-based synthesis conditioned on partial volume maps. In: Burgos, N., Svoboda, D., Wolterink, J.M., Zhao, C. (eds.) SASHIMI 2020. LNCS, vol. 12417, pp. 11–20. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59520-3_2
Segato, A., et al.: Data augmentation of 3d brain environment using deep convolutional refined auto-encoding alpha GAN. IEEE Trans. Med. Robot. Bion. 3(1), 269–272 (2020)
Kwon, G., Han, C., Kim, D.: Generation of 3D brain MRI using auto-encoding generative adversarial networks. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 118–126. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32248-9_14
Xing, S., et al.: Cycle consistent embedding of 3D brains with auto-encoding generative adversarial networks. In: Medical Imaging with Deep Learning (2021)
Sun, L., et al.: Hierarchical amortized training for memory-efficient high resolution 3D GAN. arXiv preprint arXiv:2008.01910 (2020)
Wang, Z., et al.: Multiscale structural similarity for image quality assessment. In: The Thrity-Seventh Asilomar Conference on Signals, Systems & Computers, 2003, vol. 2, pp. 1398–1402. IEEE (2003)
Heusel, M., et al.: GANs trained by a two time-scale update rule converge to a local Nash equilibrium. In: Advances in Neural Information Processing Systems, vol. 30 (2017)
Gretton, A., et al.: A kernel two-sample test. J. Mach. Learn. Res. 13(1), 723–773 (2012)
Razavi, A., et al.: Generating diverse high-fidelity images with VQ-VAE-2. In: Proceedings of the 33rd International Conference on Advances in Neural Information Processing Systems, vol. 32 (2019)
Esser, P., et al.: Taming transformers for high-resolution image synthesis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12873–12883 (2021)
Yu, J., et al.: Vector-quantized image modeling with improved VQGAN. arXiv preprint arXiv:2110.04627 (2021)
Van Den Oord, A., Vinyals, O., et al.: Neural discrete representation learning. In: Proceedings of the 31st International Conference on Advances in Neural Information Processing Systems, vol. 30 (2017)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, vol. 30 (2017)
Krzysztof, C., et al.: Rethinking attention with performers. In: Proceedings of ICLR (2021)
Jordon, J., et al.: Synthetic data-what, why and how? arXiv preprint arXiv:2205.03257 (2022)
Esteban, C., et al.: Real-valued (medical) time series generation with recurrent conditional GANs. arXiv preprint arXiv:1706.02633 (2017)
Ashburner, J., Friston, K.J.: Voxel-based morphometry-the methods. Neuroimage 11(6), 805–821 (2000)
Cardoso, M.J., et al.: Geodesic information flows: spatially-variant graphs and their application to segmentation and fusion. IEEE Trans. Med. Imaging 34(9):1976–1988 (2015)
Tay, V., et al.: Long range arena: a benchmark for efficient transformers. In: International Conference on Learning Representations (2020)
Graham, M.S., et al.: Transformer-based out-of-distribution detection for clinically safe segmentation. In: Conference on Medical Imaging with Deep Learning (2022)
Dhariwal, P., et al.: Jukebox: a generative model for music. arXiv preprint arXiv:2005.00341 (2020)
Zhang, R., et al.: The unreasonable effectiveness of deep features as a perceptual metric. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 586–595 (2018)
Isola, P., et al.: Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1125–1134 (2017)
Mao, X., et al.: Least squares generative adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2794–2802 (2017)
Tudosiu, P.-D., et al.: Neuromorphologicaly-preserving volumetric data encoding using VQ-VAE. arXiv preprint arXiv:2002.05692 (2020)
Gulrajani, I., et al.: Improved training of Wasserstein GANs. In: Conference on Advances in Neural Information Processing Systems, vol. 30 (2017)
Ridgway, G.R., et al.: The problem of low variance voxels in statistical parametric mapping; a new hat avoids a ‘haircut’. Neuroimage 59(3), 2131–2141 (2012)
Pinaya, W.H.L., et al.: Unsupervised brain anomaly detection and segmentation with transformers. In: Conference on Medical Imaging with Deep Learning, pp. 596–617. PMLR (2021)
Bachlechner, T., et al.: ReZero is all you need: Fast convergence at large depth. In: Uncertainty in Artificial Intelligence, pp. 1352–1361. PMLR (2021)
Ashburner, J., et al.: SPM12 Manual. Wellcome Trust Centre for Neuroimaging, London (2014)
Acknowledgements
WHLP, MG, PB, MJC and PN are supported by Wellcome [WT213038/Z/18/Z]. PTD is supported by the EPSRC Research Council, part of the EPSRC DTP [EP/R513064/1]. FV is supported by Wellcome/ EPSRC Centre for Medical Engineering [WT203148/Z/16/Z], Wellcome Flagship Programme [WT213038/Z/18/Z], The London AI Centre for Value-based Healthcare and GE Healthcare. PB is also supported by Wellcome Flagship Programme [WT213038/Z/18/Z] and Wellcome EPSRC CME [WT203148/Z/16/Z]. PN is also supported by the UCLH NIHR Biomedical Research Centre. The models in this work were trained on NVIDIA Cambridge-1, the UK’s largest supercomputer, aimed at accelerating digital biology.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
A 6 Appendix
A 6 Appendix
1.1 A.1 6.1 VQ-VAEs
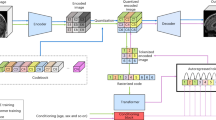
The VQ-VAE model has a similar architecture with [33] but in 3D. The encoder uses strided convolutions with stride 2 and kernel size 4. There are four downsamplings in this VQ-VAE, giving the downsampling factor \(f=2^4\). After the downsampling layers, there are three residuals blocks (\(3\times 3\times 3\) Conv, ReLU, 1\(\,\times \,\)1\(\,\times \,\)1 Conv, ReLU). The decoder mirrors the encoder and uses transposed convolutions with stride 2 and kernel size 4. All convolution layers have 256 kernels. The \(\beta \) in Eq. 1 is 0.25 and the \(\gamma \) in Eq. 2 is 0.5. The codebook size was 2048 while each element’s size was 32.
1.2 B.2 6.2 Transformers
Performer’sFootnote 1 [19] has \(L=24\) layers, \(d=256\) embedding size, 16 multi-head attention modules (8 are local attention heads with window size of 420), and ReZero gating [34]. Before the raster style ordering input was RAS+ canonical voxel representation oriented.
1.3 C.3 6.3 Losses
VQ-VAE’s pixel-space loss weight is 1.0, perceptual loss’ weight is 0.001, frequency loss’ weight is 1.0. The LPIPS uses AlexNet. Adam has been used as optimizer with an exponential decay of 0.99999. VQ-VAE’s learning rate was 0.000165, discriminator’s learning rate was 0.00005 and Performer’s CrossEntropy learning rate was 0.001.
1.4 D.4 6.4 Datasets
All datasets have been split into training and testing sub-sets. The VQ-VAE UKB sub-sets had 31740 and 3970 subjects respectively, while VQ-VAE ADNI had 648 and 82. All datasets have been first processed with a rigid body registration such that they roughly fit the same field of view. Afterwards, all samples are passed through the following transformations before being fed into the VQ-VAE during training: first, they are being normalized to [0, 1], then tightly spatially cropped resulting in an image of size (160, 224, 160), random affine (rotation range 0.04, translation range 2, scale range 0.05), random contrast adjustment (gamma [0.99, 1.01]), random intensity shift (offsets [0.0, 0.05]), random Gaussian noise (mean 0.0, standard deviation 0.02), and finally, the images were thresholded to be in the range [0, 1.0]. For the Transformer, the UKB and ADNI datasets were split into sub-populations. UKB was split into small ventricles (6388 and 108), big ventricles (6321 and 156), young (6633 and 113), old (5137 and 106), while ADNI was split into cognitively normal (118 and 29) and Alzheimer’s disease (151 and 36). For the Transformer training, each ADNI sample has been augmented 100 times and each augmentation’s index-based representation was used for training it.
1.5 E.5 6.5 VBM Analysis
For the Voxel-Based Morphometry (VBM), Statistical Parametric Mapping (SPM) [35] package version 12.7486 was used with MATLAB R2019a. Before running the statistical tests, the images must first undergo unified segmentation where they were spatially normalized to a common template and simultaneously segmented into the Gray Matter (GM), White Matter (WM), and Cerebrospinal fluid (CSF) tissue segments based on prior probability maps and voxel intensities. The unified segmentation was done with the default parameters: Bias Regularisation (light regularisation 0.001), Bias FWHM (60 mm cutoff), MRF Parameter (1), Clean Up (Light Clean), Warping Regularisation ([0, 0.001, 0.5, 0.05, 0.2]), Affine Regularisation (ICBM space template - European brains), Smoothness (0), Sampling Distance (3). As per standard practice when using VBM, the group-aligned segmentations were modulated to preserve tissue volume, and a smoothing kernel was applied to the modulated tissue compartments to make the data conform to the Gaussian field model that underlines VBM and to increase the sensitivity to detect structural changes. The smoothing was also done with the default parameters with FWHM ([8, 8, 8]). For the VBM analysis, a Two-sample t-test Design was used, with the following parameters: Independence (Yes), Variance (Unequal), Grand mean scaling (No) and ANCOVA (No). No covariates, masking or global normalisation have been used.
Appendix F - Additional Samples
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Tudosiu, PD. et al. (2022). Morphology-Preserving Autoregressive 3D Generative Modelling of the Brain. In: Zhao, C., Svoboda, D., Wolterink, J.M., Escobar, M. (eds) Simulation and Synthesis in Medical Imaging. SASHIMI 2022. Lecture Notes in Computer Science, vol 13570. Springer, Cham. https://doi.org/10.1007/978-3-031-16980-9_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-16980-9_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16979-3
Online ISBN: 978-3-031-16980-9
eBook Packages: Computer ScienceComputer Science (R0)