Abstract
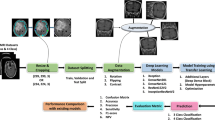
Spinal tumors contain multiple pathological subtypes, and different subtypes may correspond to different treatments and prognoses. Diagnosis of spinal tumor subtypes from medical images in the early stage is of great clinical significance. Due to the complex morphology and high heterogeneity of spinal tumors, it can be challenging to diagnose subtypes from medical images accurately. In recent years, a number of researchers have applied deep learning technology to medical image analysis. However, such research usually requires a large number of labeled samples for training, which can be difficult to obtain. Therefore, the use of unlabeled medical images to improve the identification performance of models is a hot research topic. This study proposed a self-supervised learning based pre-trained method Res-MAE using a convolutional neural network and masked autoencoder. First, this method trains an efficient feature encoder using a large amount of unlabeled spinal medical data with an image reconstruction task. Then this encoder is transferred to the downstream subtype classification in a multi-modal fusion model for fine-tuning. This multi-modal fusion model adopts a bipartite graph and multi-branch for spinal tumor subtype classification. The experimental results show that the accuracy of the proposed method can be increased by up to 10.3%, and the F1 can be increased by up to 13.8% compared with the baseline method.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Shrivastava, R.K., Epstein, F.J., Perin, N.I., et al.: Intramedullary spinal cord tumors in patients older than 50 years of age: management and outcome analysis. J. Neurosurg. Spine 2(3), 249–255 (2005)
Huang, Z., et al.: Universal semi-supervised learning. Adv. Neural Inf. Process. Syst. 34, 26714–26725 (2021)
Chowdhury, A., et al.: Applying self-supervised learning to medicine: review of the state of the art and medical implementations. In: Informatics, vol. 8, no. 3. MDPI (2021)
Liu, Q., Yu, L., Luo, L., et al.: Semi-supervised medical image classification with relation-driven self-ensembling model. IEEE Trans. Med. Imaging 39(11), 3429–3440 (2020)
Tseng, K.K., Zhang, R., Chen, C.M., et al.: DNetUNet: a semi-supervised CNN of medical image segmentation for super-computing AI service. J. Supercomput. 77(4), 3594–3615 (2021)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Valverde, J.M., et al.: Transfer learning in magnetic resonance brain imaging: a systematic review. J. Imaging 7(4), 66 (2021)
Dosovitskiy, A., Beyer, L., Kolesnikov, A., et al.: An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
Vaswani, A., et al.: Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 6000–6010 (2017)
He, K., Chen, X., Xie, S., et al.: Masked autoencoders are scalable vision learners. arXiv preprint arXiv:2111.06377 (2021)
Russakovsky, O., et al.: Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 115(3), 211–252 (2015)
Song, X., et al.: Cross-modal attention for mri and ultrasound volume registration. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12904, pp. 66–75. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87202-1_7
Zhou, T., et al.: Deep multi-modal latent representation learning for automated dementia diagnosis. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11767, pp. 629–638. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32251-9_69
Zhang, Y., et al.: Modality-aware mutual learning for multi-modal medical image segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 589–599. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_56
Zhang, Y., et al.: Multi-phase liver tumor segmentation with spatial aggregation and uncertain region inpainting. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 68–77. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_7
Syazwany, N.S., Nam, J.-H., Lee, S.-C.: MM-BiFPN: multi-modality fusion network with Bi-FPN for MRI brain tumor segmentation. IEEE Access 9, 160708–160720 (2021)
Acknowledgement
This work was supported by the Beijing Natural Science Foundation (Z190020), National Natural Science Foundation of China (82171927, 81971578), Capital's Funds for Health Improvement and Research (2020-4-40916).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jiao, M. et al. (2022). Self-supervised Learning Based on a Pre-trained Method for the Subtype Classification of Spinal Tumors. In: Qin, W., Zaki, N., Zhang, F., Wu, J., Yang, F. (eds) Computational Mathematics Modeling in Cancer Analysis. CMMCA 2022. Lecture Notes in Computer Science, vol 13574. Springer, Cham. https://doi.org/10.1007/978-3-031-17266-3_6
Download citation
DOI: https://doi.org/10.1007/978-3-031-17266-3_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-17265-6
Online ISBN: 978-3-031-17266-3
eBook Packages: Computer ScienceComputer Science (R0)