Abstract
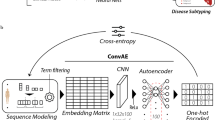
The study of existing links among different types of medical concepts can support research on optimal pathways for the treatment of human diseases. Here, we present a clustering analysis of medical concept learned representations generated from MIMIC-IV, an open dataset of de-identified digital health records. Patient’s trajectory information were extracted in chronological order to generate +500k sequence-like data structures, which were fed to a word2vec model to automatically learn concept representations. As a result, we obtained concept embeddings that describe diagnostics, procedures, and medications in a continuous low-dimensional space. A quantitative evaluation of the embeddings shows the significant power of the extracted embeddings on predicting exact labels of diagnoses, procedures, and medications for a given patient trajectory, achieving top-10 and top-30 accuracy over 47% and 66%, respectively, for all the dimensions evaluated. Moreover, clustering analyses of medical concepts after dimensionality reduction with t-SNE and UMAP techniques show that similar diagnoses (and procedures) are grouped together matching the categories of ICD-10 codes. However, the distribution by categories is not as evident if PCA or SVD are employed, indicating that the relationships among concepts are highly non-linear. This highlights the importance of non-linear models, such as those provided by deep learning, to capture the complex relationships of medical concepts.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Choi, E., Bahadori, M.T., Schuetz, A., Stewart, W.F., Sun, J.: Doctor AI: Predicting Clinical Events via Recurrent Neural Networks (2016). https://proceedings.mlr.press/v56/Choi16.html
De Freitas, J.K., et al.: Phe2vec: automated disease phenotyping based on unsupervised embeddings from electronic health records. Patterns 2(9), 100337 (2021). https://doi.org/10.1016/j.patter.2021.100337
Flamholz, Z.N., Crane-Droesch, A., Ungar, L.H., Weissman, G.E.: Word embeddings trained on published case reports are lightweight, effective for clinical tasks, and free of protected health information. J. Biomed. Inform. 125, 103971 (2022). https://doi.org/10.1016/j.jbi.2021.103971. https://www.sciencedirect.com/science/article/pii/S1532046421003002
Glynn, E.F., Hoffman, M.A.: Heterogeneity introduced by EHR system implementation in a de-identified data resource from 100 non-affiliated organizations. JAMIA Open 2(4), 554–561 (2019). https://doi.org/10.1093/jamiaopen/ooz035. https://pubmed.ncbi.nlm.nih.gov/32025653
Hartigan, J.A., Wong, M.A.: Algorithm AS 136: a K-means clustering algorithm. J. R. Stat. Soc. C: Appl. Stat. 28(1), 100–108 (1979). https://doi.org/10.2307/2346830. Full publication date 1979
Hua, R., Liu, X., Yuan, E.: Red blood cell distribution width at admission predicts outcome in critically ill patients with kidney failure: a retrospective cohort study based on the MIMIC-IV database. Ren. Fail. 44(1), 1182–1191 (2022). https://doi.org/10.1080/0886022X.2022.2098766. pMID: 35834358
Johnson, A.E., Bulgarelli, L., Pollard, T.J., Horng, S., Celi, L., Mark, R.G.: MIMIC-IV (version 1.0) (2021). https://doi.org/10.13026/s6n6-xd98
Landauer, T.K., Foltz, P.W., Laham, D.: An introduction to latent semantic analysis. Discourse Process. 25(2–3), 259–284 (1998). https://doi.org/10.1080/01638539809545028
Li, Z., Roberts, K., Jiang, X., Long, Q.: Distributed learning from multiple EHR databases: contextual embedding models for medical events. J. Biomed. Inform. 92, 103138 (2019). https://doi.org/10.1016/j.jbi.2019.103138. https://www.sciencedirect.com/science/article/pii/S1532046419300565
van der Maaten, L., Hinton, G.: Visualizing data using t-SNE. J. Mach. Learn. Res. 9(86), 2579–2605 (2008). http://jmlr.org/papers/v9/vandermaaten08a.html
McInnes, L., Healy, J., Saul, N., Großberger, L.: UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3(29), 861 (2018). https://doi.org/10.21105/joss.00861
Meng, C., Trinh, L., Xu, N., Enouen, J., Liu, Y.: Interpretability and fairness evaluation of deep learning models on MIMIC-IV dataset. Sci. Rep. 12(1), 7166 (2022). https://doi.org/10.1038/s41598-022-11012-2
Mikolov, T., Sutskever, I., Chen, K., Corrado, G.S., Dean, J.: Distributed representations of words and phrases and their compositionality. In: Advances in Neural Information Processing Systems, pp. 3111–3119 (2013)
Nowroozilarki, Z., Pakbin, A., Royalty, J., Lee, D.K., Mortazavi, B.J.: Real-time mortality prediction using MIMIC-IV ICU data via boosted nonparametric hazards. In: 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI), pp. 1–4 (2021). https://doi.org/10.1109/BHI50953.2021.9508537
Rasmy, L., Xiang, Y., Xie, Z., Tao, C., Zhi, D.: Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. NPJ Digit. Med. 4(1), 86 (2021). https://doi.org/10.1038/s41746-021-00455-y
Schneider, E.T.R., et al.: BioBERTpt - a Portuguese neural language model for clinical named entity recognition. In: Proceedings of the 3rd Clinical Natural Language Processing Workshop, pp. 65–72. Association for Computational Linguistics (2020). https://doi.org/10.18653/v1/2020.clinicalnlp-1.7
Si, Y., et al.: Deep representation learning of patient data from electronic health records (EHR): a systematic review. J. Biomed. Inform. 115, 103671 (2021)
Teodoro, D., et al.: Interoperability driven integration of biomedical data sources. Stud. Health Technol. Inform. 169, 185–189 (2011). https://doi.org/10.3233/978-1-60750-806-9-185. https://www.ncbi.nlm.nih.gov/pubmed/21893739
Teodoro, D., Pasche, E., Gobeill, J., Emonet, S., Ruch, P., Lovis, C.: Building a transnational biosurveillance network using semantic web technologies: requirements, design, and preliminary evaluation. J. Med. Internet Res. 14(3), e73–e73 (2012). https://doi.org/10.2196/jmir.2043. https://pubmed.ncbi.nlm.nih.gov/22642960, 22642960[pmid]
Teodoro, D., Sundvall, E., João Junior, M., Ruch, P., Miranda Freire, S.: ORBDA: an openEHR benchmark dataset for performance assessment of electronic health record servers. PloS One 13(1), e0190028–e0190028 (2018). https://doi.org/10.1371/journal.pone.0190028. https://pubmed.ncbi.nlm.nih.gov/29293556, 29293556[pmid]
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jaume-Santero, F. et al. (2022). Cluster Analysis of Low-Dimensional Medical Concept Representations from Electronic Health Records. In: Traina, A., Wang, H., Zhang, Y., Siuly, S., Zhou, R., Chen, L. (eds) Health Information Science. HIS 2022. Lecture Notes in Computer Science, vol 13705. Springer, Cham. https://doi.org/10.1007/978-3-031-20627-6_29
Download citation
DOI: https://doi.org/10.1007/978-3-031-20627-6_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-20626-9
Online ISBN: 978-3-031-20627-6
eBook Packages: Computer ScienceComputer Science (R0)