Abstract
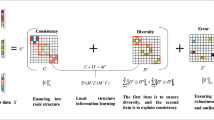
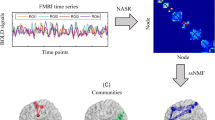
Understanding the relationship between brain function and structure is vital important in the field of brain image analysis. It elucidates the working mechanism of the brain, which will contribute to better understand the brain and simulate the brain-like system. Extensive efforts have been made on this topic, but still far from the satisfactory. The major difficulties are at least two aspects. One is the huge individual difference among the subjects, which makes it hard to obtain stable results at groupwise level, e.g., noise signals can significantly affect the exploring process. The other one is the huge difference between functional and structural features of the brain, both in their pattern and size, which are very different. To alleviate the above problems, in this paper, we propose a two-stage multi-view low-rank sparse subspace clustering (Two-stage MLRSSC) method to jointly study the relationship between brain function and structure and identify the common regions of brain function and structure. The major innovation of proposed Two-stage MLRSSC is that comparable features of brain function and structure can be effectively extracted from low-rank sparse representation, and results are further improved the stability by two-stage strategy. Finally, groupwise-based stable functional and structural common regions are identified for better understanding the relationship. Experimental results shed new ways to explore the brain function and structure, new insights are observed and discussed.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sporns, O., Tononi, G., Kötter, R.: The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, e42 (2005)
Biswal, B., Zerrin Yetkin, F., Haughton, V.M., Hyde, J.S.: Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541 (1995)
Gordon, E.M., et al.: Precision functional mapping of individual human brains. Neuron 95, 791-807.e7 (2017)
Zamora-López, G., Zhou, C., Kurths, J.: Cortical hubs form a module for multisensory integration on top of the hierarchy of cortical networks. Front. Neuroinform. 4, 1 (2010)
Sui, J., Adali, T., Yu, Q., Chen, J., Calhoun, V.D.: A review of multivariate methods for multimodal fusion of brain imaging data. J. Neurosci. Methods 204, 68–81 (2012)
Stam, C.J., et al.: The relation between structural and functional connectivity patterns in complex brain networks. Int. J. Psychophysiol. 103, 149–160 (2016)
Liang, H., Wang, H.: Structure-function network mapping and its assessment via persistent homology. PLoS Comput. Biol. 13, e1005325 (2017)
Meier, J., et al.: A mapping between structural and functional brain networks. Brain Connect. 6, 298–311 (2016)
Smith, S.M., et al.: Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 17, 666–682 (2013)
Brbić, M., Kopriva, I.: Multi-view low-rank sparse subspace clustering. Pattern Recogn. 73, 247–258 (2018)
Huntenburg, J.M., Bazin, P.-L., Margulies, D.S.: Large-scale gradients in human cortical organization. Trends Cogn. Sci. 22, 21–31 (2018)
Destrieux, C., Fischl, B., Dale, A., Halgren, E.: Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 53, 1–15 (2010)
Van Essen, D.C., et al.: The human connectome project: a data acquisition perspective. Neuroimage 62, 2222–2231 (2012)
Dale, A.M., Fischl, B., Sereno, M.I.: Cortical surface-based analysis: I segmentation and surface reconstruction. Neuroimage 9, 179–194 (1999)
Fischl, B., et al.: Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002)
Glasser, M.F., et al.: The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124 (2013)
van den Heuvel, M.P., Kahn, R.S., Goñi, J., Sporns, O.: High-cost, high-capacity backbone for global brain communication. Proc. Natl. Acad. Sci. 109, 11372–11377 (2012)
Jenkinson, M., Bannister, P., Brady, M., Smith, S.: Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002)
Jenkinson, M., Beckmann, C.F., Behrens, T.E.J., Woolrich, M.W., Smith, S.M.: FSL. Neuroimage 62, 782–790 (2012)
Zhu, D., et al.: DICCCOL: dense individualized and common connectivity-based cortical landmarks. Cereb. Cortex 23, 786–800 (2013)
Wang, Z., Bovik, A.C., Sheikh, H.R., Simoncelli, E.P.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004)
Kennedy, J., Eberhart, R.C.: A discrete binary version of the particle swarm algorithm. In: Computational Cybernetics and Simulation 1997 IEEE International Conference on Systems, Man, and Cybernetics, vol. 5, pp. 4104–4108 (1997)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, S. et al. (2022). A Novel Two-Stage Multi-view Low-Rank Sparse Subspace Clustering Approach to Explore the Relationship Between Brain Function and Structure. In: Lian, C., Cao, X., Rekik, I., Xu, X., Cui, Z. (eds) Machine Learning in Medical Imaging. MLMI 2022. Lecture Notes in Computer Science, vol 13583. Springer, Cham. https://doi.org/10.1007/978-3-031-21014-3_20
Download citation
DOI: https://doi.org/10.1007/978-3-031-21014-3_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-21013-6
Online ISBN: 978-3-031-21014-3
eBook Packages: Computer ScienceComputer Science (R0)