Abstract
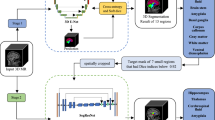
Brain parcellation plays an important role in neurodegenerative disease diagnosis and brain network analysis. One of the big challenges in brain parcellation is lack of clear anatomical boundary between different brain regions. As a result, for the task involving a large number of brain regions, i.e., during fine brain parcellation, the parcellation accuracy could be significantly degraded. Unfortunately, few studies focused on this issue. To this end, we propose a novel multi-scale deep brain parcellation network. Specifically, different scales of brain regions, i.e., global, middle and fine scales, are defined. From global to fine scales, brain regions are gradually subdivided and refined. The proposed deep network performs brain parcellation at each scale simultaneously (multi-task), where parcellation at fine scale is under the constraint of large scales. In addition, we also present a new focal region based auxiliary network, which focuses on the brain regions difficult to be parcellated at fine scale. The final parcellation results are obtained by merging the outputs of the brain parcellation backbone at all scales and the focal region based auxiliary network. Comparison and ablation experiments are conducted on a multi-center clinical brain MRI dataset of 267 subjects with 101 brain regions. Experimental results demonstrate that the proposed approach outperforms the state-of-the-art methods under comparison.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38(8), 1788–1800 (2019)
Coupé, P., et al.: AssemblyNet: a large ensemble of CNNs for 3D whole brain MRI segmentation. NeuroImage 219, 117026 (2020)
Desikan, R.S., et al.: An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31(3), 968–980 (2006)
Fischl, B.: FreeSurfer. NeuroImage 62(2), 774–781 (2012)
Henschel, L., Conjeti, S., Estrada, S., Diers, K., Fischl, B., Reuter, M.: FastSurfer-a fast and accurate deep learning based neuroimaging pipeline. NeuroImage 219, 117012 (2020)
Huo, Y., et al.: 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage 194, 105–119 (2019)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Milletari, F., Navab, N., Ahmadi, S.A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Roy, A.G., Conjeti, S., Navab, N., Wachinger, C., Initiative, A.D.N., et al.: QuickNAT: a fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage 186, 713–727 (2019)
Roy, A.G., Conjeti, S., Sheet, D., Katouzian, A., Navab, N., Wachinger, C.: Error corrective boosting for learning fully convolutional networks with limited data. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 231–239. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_27
Sun, L., Shao, W., Wang, M., Zhang, D., Liu, M.: High-order feature learning for multi-atlas based label fusion: application to brain segmentation with MRI. IEEE Trans. Image Process. 29, 2702–2713 (2020)
Sun, L., Shao, W., Zhang, D., Liu, M.: Anatomical attention guided deep networks for ROI segmentation of brain MR images. IEEE Trans. Med. Imaging 39(6), 2000–2012 (2020)
Wang, H., Suh, J.W., Das, S.R., Pluta, J.B., Craige, C., Yushkevich, P.A.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 611–623 (2013)
Wu, Z., Guo, Y., Park, S.H., Gao, Y., Dong, P., Lee, S.W., Shen, D.: Robust brain ROI segmentation by deformation regression and deformable shape model. Med. Image Anal. 43, 198–213 (2018)
Xiao, X., Lian, S., Luo, Z., Li, S.: Weighted Res-UNet for high-quality retina vessel segmentation. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327–331. IEEE (2018)
Zuo, X.N., et al.: An open science resource for establishing reliability and reproducibility in functional connectomics. Sci. Data 1(1), 1–13 (2014)
Acknowledgement
This work was supported in part by National Key Research and Development Program of China (Grant No. 2017YFA0700800), National Natural Science Foundation of China (Grant No. 62131015, 62088102 and 62073012), Science and Technology Commission of Shanghai Municipality (STCSM) (Grant No. 21010502600), Natural Science Basic Research Plan in Shaanxi Province of China (Grant No. 2022JM-324), Key Research and Development Program of Shaanxi Province of China (Grant No. 2020GXLHY-008), Social Science Foundation of Shaanxi Province of China (Grant No. 2021K014), and Beijing Municipal Natural Science Foundation (Grant No. 7222307).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Ge, Y. et al. (2022). Multi-scale and Focal Region Based Deep Learning Network for Fine Brain Parcellation. In: Lian, C., Cao, X., Rekik, I., Xu, X., Cui, Z. (eds) Machine Learning in Medical Imaging. MLMI 2022. Lecture Notes in Computer Science, vol 13583. Springer, Cham. https://doi.org/10.1007/978-3-031-21014-3_48
Download citation
DOI: https://doi.org/10.1007/978-3-031-21014-3_48
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-21013-6
Online ISBN: 978-3-031-21014-3
eBook Packages: Computer ScienceComputer Science (R0)