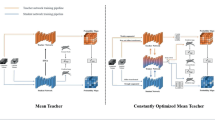
Abstract
Accurate segmentation of anatomical structures is a critical step for medical image analysis. Deep learning architectures have become the state-of-the-art models for automatic medical image segmentation. However, these models require an extensive labelled dataset to achieve a high performance. Given that obtaining annotated medical datasets is very expensive, in this work we present a two-phase teacher-student approach for semi-supervised learning. In phase 1, a three network U-Net ensemble, denominated the teacher, is trained using the labelled dataset. In phase 2, a student U-Net network is trained with the labelled dataset and the unlabelled dataset with pseudo-labels produced with the teacher network. The student network is then used for inference of the testing images. The proposed approach is evaluated on the task of abdominal segmentation from the FLARE2022 challenge, achieving a mean 0.53 dice, 0.57 NSD, and 44.97 prediction time on the validation set.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Baldeon Calisto, M., Lai-Yuen, S.K.: AdaEn-Net : an ensemble of adaptive 2D–3D fully convolutional networks for medical image segmentation. Neural Netw. 126, 76–94 (2020). https://doi.org/10.1016/j.neunet.2020.03.007
Baldeon-Calisto, M., Lai-Yuen, S.K.: AdaResU-Net: Multiobjective adaptive convolutional neural network for medical image segmentation. Neurocomputing 392, 325–340 (2020). https://doi.org/10.1016/j.neucom.2019.01.110
Buslaev, A., Iglovikov, V.I., Khvedchenya, E., Parinov, A., Druzhinin, M., Kalinin, A.A.: Albumentations: fast and flexible image augmentations. Information 11(2) (2020). https://doi.org/10.3390/info11020125, https://www.mdpi.com/2078-2489/11/2/125
Chen, S., Bortsova, G., García-Uceda Juárez, A., van Tulder, G., de Bruijne, M.: Multi-task attention-based semi-supervised learning for medical image segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 457–465. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32248-9_51
Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., et al.: The cancer imaging archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26(6), 1045–1057 (2013)
Heller, N., et al.: The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: results of the kits19 challenge. Med. Image Anal. 67, 101821 (2021)
Heller, N., et al.: An international challenge to use artificial intelligence to define the state-of-the-art in kidney and kidney tumor segmentation in CT imaging. Proc. Am. Soc. Clin. Oncol. 38(6), 626–626 (2020)
Lee, C.Y., Xie, S., Gallagher, P., Zhang, Z., Tu, Z.: Deeply-supervised nets. In: Proceedings of the Eighteenth International Conference on Artificial Intelligence and Statistics, PMLR 38 2015. pp. 562–570 (2015)
Luo, X., Chen, J., Song, T., Wang, G.: Semi-supervised medical image segmentation through dual-task consistency. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 35, pp. 8801–8809 (2021)
Ma, J., et al.: Loss odyssey in medical image segmentation. Med. Image Anal. 71, 102035 (2021)
Ma, J., et al.: Fast and low-GPU-memory abdomen CT organ segmentation: the flare challenge. Med. Image Anal. 82, 102616 (2022). https://doi.org/10.1016/j.media.2022.102616
Ma, J., et al.: AbdomenCT-1K: Is abdominal organ segmentation a solved problem? IEEE Trans. Pattern Anal. Mach. Intell. 44(10), 6695–6714 (2022)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241 (2015)
Simpson, A.L., et al.: A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 (2019)
Tomar, D., Lortkipanidze, M., Vray, G., Bozorgtabar, B., Thiran, J.P.: Self-attentive spatial adaptive normalization for cross-modality domain adaptation. IEEE Trans. Med. Imaging 40(10), 2926–2938 (2021)
Van Engelen, J.E., Hoos, H.H.: A survey on semi-supervised learning. Mach. Learn. 109(2), 373–440 (2020)
Acknowledgements
The authors of this paper declare that the segmentation method they implemented for participation in the FLARE 2022 challenge has not used any pre-trained models nor additional datasets other than those provided by the organizers.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Baldeon Calisto, M. (2022). Teacher-Student Semi-supervised Approach for Medical Image Segmentation. In: Ma, J., Wang, B. (eds) Fast and Low-Resource Semi-supervised Abdominal Organ Segmentation. FLARE 2022. Lecture Notes in Computer Science, vol 13816. Springer, Cham. https://doi.org/10.1007/978-3-031-23911-3_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-23911-3_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-23910-6
Online ISBN: 978-3-031-23911-3
eBook Packages: Computer ScienceComputer Science (R0)