Abstract
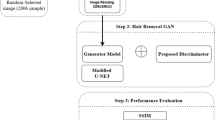
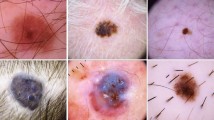
Dermoscopic images are often contaminated by artifacts including clinical pen markings, immersion fluid air bubbles, dark corners, and most importantly hair, which makes interpreting them more challenging for clinicians and computer-aided diagnostic algorithms. Hence, automated artifact recognition and inpainting systems have the potential to aid the clinical workflow as well as serve as an preprocessing step in the automated classification of dermoscopic images. In this paper, we share the first release of a public dermoscopic image dataset with hair artifacts which can be accessed here https://skin-hairdataset.github.io/SHD/. The Skin_Hair dataset contains over 252 dermoscopic images including artificial hair and will be expanded over time. Furthermore, we present the primary results of applying machine learning algorithms and GAN based architectures to the hair inpainting problem in dermoscopic images. We envision that these results will serve as a benchmark for researchers who might work on the hair detection and reconstruction tasks with this dataset in the future. In this work, we present a skin lesion image dataset based on the ISIC dataset containing dermoscopic images, images containing artificial hairs and the corresponding ground-truth masks. Furthermore, we use four hair inpainting methods including Navier-Stokes, Telea, Hair_SinGAN and R-MNet architectures which we evaluate using image quality assessment metrics MSE, PSNR, UQI and SSIM. The R-MNet architecture achieved the highest SSIM score of 0.960.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abbas, Q., Celebi, M., García, I.F.: Hair removal methods: a comparative study for dermoscopy images. Biomed. Signal Process. Control 6(4), 395–404 (2011). https://doi.org/10.1016/j.bspc.2011.01.003, https://www.sciencedirect.com/science/article/pii/S1746809411000048
Adegun, A.A., Viriri, S.: Deep learning-based system for automatic melanoma detection. IEEE Access 8, 7160–7172 (2020)
Adegun, A.A., Viriri, S.: Deep learning techniques for skin lesion analysis and melanoma cancer detection: a survey of state-of-the-art. Artif. Intell. Rev. 54, 811–841 (2020)
Almaraz-Damian, J.A., Ponomaryov, V., Sadovnychiy, S., Castillejos-Fernandez, H.: Melanoma and nevus skin lesion classification using handcraft and deep learning feature fusion via mutual information measures. Entropy 22(4) (2020). https://doi.org/10.3390/e22040484, https://www.mdpi.com/1099-4300/22/4/484
Barbosa, J., Baleiras, M.: Melanoma detection using deep learning methods (2019)
Bardou, D., Bouaziz, H., Lv, L., Zhang, T.: Hair removal in dermoscopy images using variational autoencoders. Skin Res. Technol. 28(3), 445–454 (2022). https://doi.org/10.1111/srt.13145, https://onlinelibrary.wiley.com/doi/abs/10.1111/srt.13145
Bertalmio, M., Bertozzi, A.L., Sapiro, G.: Navier-stokes, fluid dynamics, and image and video inpainting. In: Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. CVPR 2001, vol. 1, pp. I-I. IEEE (2001)
Bisla, D., Choromanska, A., Berman, R., Stein, J., Polsky, D.: Towards automated melanoma detection with deep learning: data purification and augmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), pp. 2720–2728 (2019)
Bisla, D., Choromanska, A., Stein, J., Polsky, D., Berman, R.: Skin lesion segmentation and classification with deep learning system. arXiv:abs/1902.06061 (2019)
Borys, D., Kowalska, P., Frackiewicz, M., Ostrowski, Z.: A simple hair removal algorithm from dermoscopic images. In: Ortuño, F., Rojas, I. (eds.) IWBBIO 2015. LNCS, vol. 9043, pp. 262–273. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-16483-0_27
Cassidy, B., Kendrick, C., Brodzicki, A., Jaworek-Korjakowska, J., Yap, M.H.: Analysis of the ISIC image datasets: usage, benchmarks and recommendations. Med. Image Anal. 75, 102305 (2022)
Celebi, M.E., Barata, C., Halpern, A., Tschandl, P., Combalia, M., Liu, Y.: Guest editorial: image analysis in dermatology. Med. Image Anal. 79, 102468 (2022). https://doi.org/10.1016/j.media.2022.102468
Chan, T.F., Shen, J.: Nontexture inpainting by curvature-driven diffusions. J. Visual Commun. Image Represent. 12(4), 436–449 (2001). https://doi.org/10.1006/jvci.2001.0487, https://www.sciencedirect.com/science/article/pii/S1047320301904870
Codella, N., et al.: Skin lesion analysis toward melanoma detection 2018: a challenge hosted by the international skin imaging collaboration (ISIC) (2018)
Codella, N.C., et al.: Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 168–172. IEEE (2018)
Combalia, M., et al.: Bcn20000: Dermoscopic lesions in the wild (2019)
Fiorese, M., Peserico, E., Silletti, A.: Virtualshave: automated hair removal from digital dermatoscopic images. In: 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 5145–5148 (2011). https://doi.org/10.1109/IEMBS.2011.6091274
Gutman, D., et al.: Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, Hosted by the International Skin Imaging Collaboration (ISIC) (2016)
Horé, A., Ziou, D.: Image quality metrics: PSNR vs. SSIM. In: 2010 20th International Conference on Pattern Recognition, pp. 2366–2369 (2010). https://doi.org/10.1109/ICPR.2010.579
Huang, A., Kwan, S.Y., Chang, W.Y., Liu, M.Y., Chi, M.H., Chen, G.S.: A robust hair segmentation and removal approach for clinical images of skin lesions. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 3315–3318 (2013). https://doi.org/10.1109/EMBC.2013.6610250
ISIC: Isic archive gallery. Online, July 2020. https://www.isic-archive.com
Jam, J., Kendrick, C., Drouard, V., Walker, K., Hsu, G.S., Yap, M.H.: R-MNET: a perceptual adversarial network for image inpainting. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 2714–2723 (2021)
Kassem, M.A., Hosny, K.M., Fouad, M.M.: Skin lesions classification into eight classes for ISIC 2019 using deep convolutional neural network and transfer learning. IEEE Access 8, 114822–114832 (2020). https://doi.org/10.1109/ACCESS.2020.3003890
Kiani, K., Sharafat, A.R.: E-shaver: an improved dullrazor® for digitally removing dark and light-colored hairs in dermoscopic images. Comput. Biol. Med. 41(3), 139–145 (2011). https://doi.org/10.1016/j.compbiomed.2011.01.003, https://www.sciencedirect.com/science/article/pii/S0010482511000047
Lee, T., Ng, V., Gallagher, R., Coldman, A., McLean, D.: Dullrazor®: A software approach to hair removal from images. Comput. Biol. Med. 27(6), 533–543 (1997). https://doi.org/10.1016/S0010-4825(97)00020-6, https://www.sciencedirect.com/science/article/pii/S0010482597000206
Li, W., Joseph Raj, A.N., Tjahjadi, T., Zhuang, Z.: Digital hair removal by deep learning for skin lesion segmentation. Pattern Recogn. 117, 107994 (2021). https://doi.org/10.1016/j.patcog.2021.107994, https://www.sciencedirect.com/science/article/pii/S0031320321001813
Maglogiannis, I., Delibasis, K.: Hair removal on dermoscopy images. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 2960–2963 (2015). https://doi.org/10.1109/EMBC.2015.7319013
Maron, R.C., et al.: Reducing the impact of confounding factors on skin cancer classification via image segmentation: technical model study. J. Med. Internet Res. 23 (2021)
Nauta, M., Walsh, R., Dubowski, A., Seifert, C.: Uncovering and correcting shortcut learning in machine learning models for skin cancer diagnosis. Diagnostics 12(1) (2022). https://doi.org/10.3390/diagnostics12010040, https://www.mdpi.com/2075-4418/12/1/40
Pewton, S.W., Yap, M.H.: Dark corner on skin lesion image dataset: does it matter? In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, pp. 4831–4839, June 2022
Rotemberg, V., et al.: A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 8, 34 (2021). https://doi.org/10.1038/s41597-021-00815-z
Rott Shaham, T., Dekel, T., Michaeli, T.: SinGAN: Learning a generative model from a single natural image. In: IEEE International Conference on Computer Vision (ICCV)(2019)
Salido, J.A.A., Ruiz, C.: Using morphological operators and inpainting for hair removal in dermoscopic images. In: Proceedings of the Computer Graphics International Conference. CGI 2017, Association for Computing Machinery, New York, NY, USA (2017). https://doi.org/10.1145/3095140.3095142, https://doi.org/10.1145/3095140.3095142
Sethian, J.A.: A fast marching level set method for monotonically advancing fronts. Proc. Natl. Acad. Sci. 93(4), 1591–1595 (1996)
Shaham, T.R., Dekel, T., Michaeli, T.: SinGAN: learning a generative model from a single natural image. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 4570–4580 (2019)
Song, X., et al.: Research on hair removal algorithm of dermatoscopic images based on maximum variance fuzzy clustering and optimization criminisi algorithm. Biomed. Signal Process. Control 78, 103967 (2022). https://doi.org/10.1016/j.bspc.2022.103967, https://www.sciencedirect.com/science/article/pii/S1746809422004669
Sultana, A., Dumitrache, I., Vocurek, M., Ciuc, M.: Removal of artifacts from dermatoscopic images. In: 2014 10th International Conference on Communications (COMM), pp. 1–4 (2014). https://doi.org/10.1109/ICComm.2014.6866757
Talavera-Martínez, L., Bibiloni, P., González-Hidalgo, M.: Comparative study of dermoscopic hair removal methods. In: Tavares, J.M.R.S., Natal Jorge, R.M. (eds.) VipIMAGE 2019. LNCVB, vol. 34, pp. 12–21. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32040-9_2
Telea, A.: An image inpainting technique based on the fast marching method. J. Graphics Tools 9(1), 23–34 (2004)
Toossi, M.T.B., Pourreza, H.R., Zare, H., Sigari, M.H., Layegh, P., Azimi, A.: An effective hair removal algorithm for dermoscopy images. Skin Res. Technol. 19 (2013)
Tschandl, P.: The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions (2018). https://doi.org/10.7910/DVN/DBW86T, https://doi.org/10.7910/DVN/DBW86T
Wang, Z., Bovik, A.C.: Mean squared error: love it or leave it? A new look at signal fidelity measures. IEEE Signal Process. Mag. 26(1), 98–117 (2009). https://doi.org/10.1109/MSP.2008.930649
Xie, F.Y., Qin, S.Y., Jiang, Z.G., Meng, R.S.: PDE-based unsupervised repair of hair-occluded information in dermoscopy images of melanoma. Comput. Med. Imaging Graph. 33(4), 275–282 (2009). https://doi.org/10.1016/j.compmedimag.2009.01.003, https://www.sciencedirect.com/science/article/pii/S0895611109000056
Xie, Y., Zhang, J., Xia, Y., Shen, C.: A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Trans. Med. Imaging 39, 2482–2493 (2020)
Zanddizari, H., Nguyen, N., Zeinali, B., Chang, J.M.: A new preprocessing approach to improve the performance of CNN-based skin lesion classification. Med. Biol. Eng. Comput. 59(5), 1123–1131 (2021). https://doi.org/10.1007/s11517-021-02355-5
Wang, Z., Bovik, A.C., Sheikh, H.R., Simoncelli, E.P.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13(4), 600–612 (2004). https://doi.org/10.1109/TIP.2003.819861
Acknowledgments
We gratefully acknowledge the funding support of the research project by the program “Excellence initiative—research university” for the AGH UST and the NAWA Bekker Scholarship for J. Jaworek-Korjakowska.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jaworek-Korjakowska, J. et al. (2023). Skin_Hair Dataset: Setting the Benchmark for Effective Hair Inpainting Methods for Improving the Image Quality of Dermoscopic Images. In: Karlinsky, L., Michaeli, T., Nishino, K. (eds) Computer Vision – ECCV 2022 Workshops. ECCV 2022. Lecture Notes in Computer Science, vol 13804. Springer, Cham. https://doi.org/10.1007/978-3-031-25069-9_12
Download citation
DOI: https://doi.org/10.1007/978-3-031-25069-9_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-25068-2
Online ISBN: 978-3-031-25069-9
eBook Packages: Computer ScienceComputer Science (R0)