Abstract
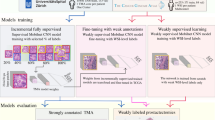
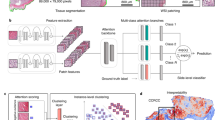
In computational pathology, predictive models from Whole Slide Images (WSI) mostly rely on Multiple Instance Learning (MIL), where the WSI are represented as a bag of tiles, each of which is encoded by a Neural Network (NN). Slide-level predictions are then achieved by building models on the agglomeration of these tile encodings. The tile encoding strategy thus plays a key role for such models. Current approaches include the use of encodings trained on unrelated data sources, full supervision or self-supervision. While self-supervised learning (SSL) exploits unlabeled data, it often requires large computational resources to train. On the other end of the spectrum, fully-supervised methods make use of valuable prior knowledge about the data but involve a costly amount of expert time. This paper proposes a framework to reconcile SSL and full supervision, showing that a combination of both provides efficient encodings, both in terms of performance and in terms of biological interpretability. On a recently organized challenge on grading Cervical Biopsies, we show that our mixed supervision scheme reaches high performance (weighted accuracy (WA): 0.945), outperforming both SSL (WA: 0.927) and transfer learning from ImageNet (WA: 0.877). We further shed light upon the internal representations that trigger classification results, providing a method to reveal relevant phenotypic patterns for grading cervical biopsies. We expect that the combination of full and self-supervision is an interesting strategy for many tasks in computational pathology and will be widely adopted by the field.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bera, K., Schalper, K.A., Rimm, D.L., Velcheti, V., Madabhushi, A.: Artificial intelligence in digital pathology-new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16(11), 703–715 (2019)
Campanella, G., et al.: Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25(8), 1301–1309 (2019). https://doi.org/10.1038/s41591-019-0508-1. https://www.nature.com/articles/s41591-019-0508-1
Chen, T., Kornblith, S., Norouzi, M., Hinton, G.: A simple framework for contrastive learning of visual representations (2020). https://arxiv.org/abs/2002.05709v3
Chung, Y.A., Lin, H.T., Yang, S.W.: Cost-aware pre-training for multiclass cost-sensitive deep learning. IJCAI (2016)
Coudray, N., et al.: Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24(10), 1559–1567 (2018). https://doi.org/10.1038/s41591-018-0177-5. https://www.nature.com/articles/s41591-018-0177-5
Courtiol, P., Tramel, E.W., Sanselme, M., Wainrib, G.: Classification and disease localization in histopathology using only global labels: a weakly-supervised approach. arXiv:1802.02212 [cs, stat] (2020)
Dehaene, O., Camara, A., Moindrot, O., de Lavergne, A., Courtiol, P.: Self-supervision closes the gap between weak and strong supervision in histology (2020). https://arxiv.org/abs/2012.03583v1
Diao, J.A., et al.: Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun. 12(1), 1613 (2021). https://doi.org/10.1038/s41467-021-21896-9. https://www.nature.com/articles/s41467-021-21896-9
DrivenData: TissueNet: Detect Lesions in Cervical Biopsies. https://www.drivendata.org/competitions/67/competition-cervical-biopsy/page/254/
Ehteshami Bejnordi, B., et al.: Consortium: diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22), 2199–2210 (2017). https://doi.org/10.1001/jama.2017.14585
Erhan, D., Bengio, Y., Courville, A., Vincent, P.: Visualizing higher-layer features of a deep network. Technical report, Univeristé de Montréal (2009)
Huang, G., Liu, Z., van der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. Technical report (2016). https://ui.adsabs.harvard.edu/abs/2016arXiv160806993H
Huang, Y.J., et al.: Rectifying supporting regions with mixed and active supervision for rib fracture recognition. IEEE Trans. Med. Imaging 39(12), 3843–3854 (2020). https://doi.org/10.1109/TMI.2020.3006138
Ilse, M., Tomczak, J.M., Welling, M.: Attention-based deep multiple instance learning (2018). https://arxiv.org/abs/1802.04712v4
Kather, J.N., et al.: Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1(8), 789–799 (2020). https://doi.org/10.1038/s43018-020-0087-6. https://www.nature.com/articles/s43018-020-0087-6
Lazard, T., et al.: Deep learning identifies morphological patterns of homologous recombination deficiency in luminal breast cancers from whole slide images. Cell Rep. Med. 3(12), 100872 (2022). https://doi.org/10.1016/j.xcrm.2022.100872
Li, J., et al.: Hybrid supervision learning for pathology whole slide image classification. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12908, pp. 309–318. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87237-3_30
Li, Z., et al.: Thoracic disease identification and localization with limited supervision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2018)
Lu, M.Y., Williamson, D.F.K., Chen, T.Y., Chen, R.J., Barbieri, M., Mahmood, F.: Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5(6), 555–570 (2021). https://doi.org/10.1038/s41551-020-00682-w. https://www.nature.com/articles/s41551-020-00682-w
Mlynarski, P., Delingette, H., Criminisi, A., Ayache, N.: Deep learning with mixed supervision for brain tumor segmentation. J. Med. Imaging 6(3), 034002 (2019). https://doi.org/10.1117/1.JMI.6.3.034002. https://www.spiedigitallibrary.org/journals/journal-of-medical-imaging/volume-6/issue-3/034002/Deep-learning-with-mixed-supervision-for-brain-tumor-segmentation/10.1117/1.JMI.6.3.034002.full
Nguyen, A., Yosinski, J., Clune, J.: Understanding neural networks via feature visualization: a survey. arXiv:1904.08939 [cs, stat] (2019)
Ruifrok, A.C., Johnston, D.A.: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23(4), 291–299 (2001)
Saillard, C., et al.: Identification of pancreatic adenocarcinoma molecular subtypes on histology slides using deep learning models. J. Clin. Oncol. 39(15_Suppl.), 4141 (2021). https://doi.org/10.1200/JCO.2021.39.15suppl.4141. https://ascopubs.org/doi/abs/10.1200/JCO.2021.39.15suppl.4141
Tourniaire, P., Ilie, M., Hofman, P., Ayache, N., Delingette, H.: Attention-based multiple instance learning with mixed supervision on the camelyon16 dataset. In: Proceedings of the MICCAI Workshop on Computational Pathology, pp. 216–226. PMLR (2021). https://proceedings.mlr.press/v156/tourniaire21a.html
Tu, H.H., Lin, H.T.: One-sided support vector regression for multiclass cost-sensitive classification, p. 8 (2010)
Weitz, P., et al.: Transcriptome-wide prediction of prostate cancer gene expression from histopathology images using co-expression based convolutional neural networks. arXiv preprint arXiv:2104.09310 (2021)
WHO: Colposcopy and treatment of cervical intraepithelial neoplasia: a beginners’ manual (2020). https://screening.iarc.fr/colpochap.php?chap=2
Acknowledgments
The authors thank Etienne Decencière for the thoughful discussions that help the project. ML was supported by a CIFRE PhD fellowship founded by KEEN EYE and ANRT (CIFRE 2019/1905). TL was supported by a Q-Life PhD fellowship (Q-life ANR-17-CONV-0005). This work was supported by the French government under management of ANR as part of the “Investissements d’avenir” program, reference ANR-19-P3IA-0001 (PRAIRIE 3IA Institute).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Lubrano, M. et al. (2023). Automatic Grading of Cervical Biopsies by Combining Full and Self-supervision. In: Karlinsky, L., Michaeli, T., Nishino, K. (eds) Computer Vision – ECCV 2022 Workshops. ECCV 2022. Lecture Notes in Computer Science, vol 13807. Springer, Cham. https://doi.org/10.1007/978-3-031-25082-8_27
Download citation
DOI: https://doi.org/10.1007/978-3-031-25082-8_27
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-25081-1
Online ISBN: 978-3-031-25082-8
eBook Packages: Computer ScienceComputer Science (R0)