Abstract
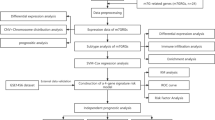
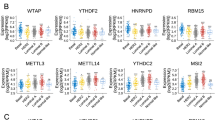
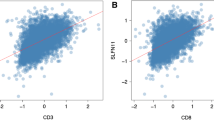
Complex signatures expressed at the genetic, transcriptional, and epigenetic levels influence tumorigenesis and evolution in breast cancer. A growing body of evidence supports the close association of RNA modification with the epigenetic regulation of the immune response. However, the mechanism of RNA modification “writers” in the immunity of breast cancer remains indeterminacy. We analyzed genomic alterations in 8236 breast cancer samples from the cBio portal database. Correlations between RNA “writers” and the expression of immunomodulators, including immunosuppressants, immunostimulants, and MHC molecules, were calculated using the TIMER and TISIDB databases. Our analysis confirmed that abnormalities in four classes of RNA “writers” were significantly correlated with poor prognosis in breast cancer. In addition, abnormal expression of TRMT61B, a tumor-associated RNA “writer”, may be associated with patients prognosis, immune infiltration levels, and expression of immunomodulators in breast cancer patients. Our results suggest that TRMT61B may serve as a biological marker of breast cancer prognosis and a potential drug target, providing a novel idea for the future therapy of breast cancer.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sung, H., et al.: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021)
Quail, D.F., Joyce, J.A.: Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013)
Lian, H., Wang, Q.-H., Zhu, C.-B., Ma, J., Jin, W.-L.: Deciphering the epitranscriptome in cancer. Trends Cancer 4, 207–221 (2018)
Huang, Z., et al.: Prognostic significance and tumor immune microenvironment heterogenicity of m5C RNA methylation regulators in triple-negative breast cancer. Front. Cell Dev. Biol. 9, 919 (2021)
Li, B., et al.: Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17, 1–16 (2016)
Cai, C., et al.: M6A “Writer” gene METTL14: a favorable prognostic biomarker and correlated with immune infiltrates in rectal cancer. Front. Oncol. 11, 2224 (2021)
Li, X., Ma, S., Deng, Y., Yi, P., Yu, J.: Targeting the RNA m6A modification for cancer immunotherapy. Mol. Cancer 21, 1–16 (2022)
Harao, M., et al.: 4-1BB–enhanced expansion of CD8+ TIL from triple-negative breast cancer unveils mutation-specific CD8+ T cells. Cancer Immunol. Res. 5, 439–445 (2017)
Deepak, K.G.K., et al.: Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 153, 104683 (2020)
Kumari, K., Groza, P., Aguilo, F.: Regulatory roles of RNA modifications in breast cancer. NAR Cancer 3, zcab036 (2021)
Boccaletto, P., et al.: MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018)
Dominissini, D., et al.: Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012)
Zhang, C., et al.: m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017)
Liu, N., Zhou, K.I., Parisien, M., Dai, Q., Diatchenko, L., Pan, T.: N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017)
Li, Y., et al.: Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol. Cancer 18, 1–6 (2019)
Liu, L., et al.: N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J. Cancer 10, 5447 (2019)
Woo, H.-H., Chambers, S.K.: Human ALKBH3-induced m1A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 35–46 (2019)
Dominissini, D., et al.: The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016)
Griseri, P., Pagès, G.: Regulation of the mRNA half-life in breast cancer. World J. Clin. Oncol. 5, 323 (2014)
Di Giammartino, D.C., Nishida, K., Manley, J.L.: Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866 (2011)
Fumagalli, D., et al.: Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 13, 277–289 (2015)
Li, T.: TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110 (2017). https://doi.org/10.1158/0008-5472
Ru, B., et al.: TISIDB: an integrated repository portal for tumor–immune system interactions. Bioinformatics 35, 4200–4202 (2019)
Shi, X., et al.: Next-generation sequencing identifies novel genes with rare variants in total anomalous pulmonary venous connection. EBioMedicine 38, 217–227 (2018)
Noh, J.H., Kim, K.M., McClusky, W.G., Abdelmohsen, K., Gorospe, M.: Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 9, e1471 (2018)
Zaccara, S., Ries, R.J., Jaffrey, S.R.: Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019)
Frye, M., Harada, B.T., Behm, M., He, C.: RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018)
Qu, Y., et al.: RNA modification “writer”-mediated RNA modification patterns and tumor microenvironment characteristics of cervical cancer. Clin. Transl. Oncol. 24, 1413–1424 (2022). https://doi.org/10.1007/s12094-022-02787-x
Chen, H., et al.: Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol. Cancer 20, 29 (2021)
Zhang, J., et al.: To develop and validate the combination of RNA methylation regulators for the prognosis of patients with gastric cancer. OncoTargets Ther. 13, 10785 (2020)
Zhao, Q., et al.: m6A RNA modification modulates PI3K/Akt/mTOR signal pathway in gastrointestinal cancer. Theranostics 10, 9528 (2020)
Zhao, Y., et al.: m1A regulated genes modulate PI3K/AKT/mTOR and ErbB pathways in gastrointestinal cancer. Transl. Oncol. 12, 1323–1333 (2019)
Jiang, X., et al.: The role of m6A modification in the biological functions and diseases. Signal Transduct. Target Ther. 6, 1–16 (2021)
Liu, T., Li, C., Jin, L., Li, C., Wang, L.: The prognostic value of m6A RNA methylation regulators in colon adenocarcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 25, 9435 (2019)
Qu, N., et al.: Multiple m6A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer 20, 1–14 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wu, P., Zhou, Y., Luo, W., Wu, L. (2023). The Potential Role of RNA “Writer” TRMT61B in the Immune Regulation of Breast Cancer. In: Wen, S., Yang, C. (eds) Biomedical and Computational Biology. BECB 2022. Lecture Notes in Computer Science(), vol 13637. Springer, Cham. https://doi.org/10.1007/978-3-031-25191-7_3
Download citation
DOI: https://doi.org/10.1007/978-3-031-25191-7_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-25190-0
Online ISBN: 978-3-031-25191-7
eBook Packages: Computer ScienceComputer Science (R0)