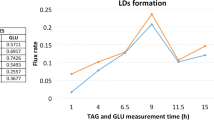
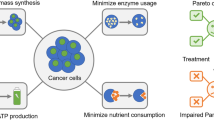
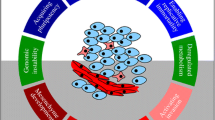
Abstract
We will consider genome-scale metabolic models that attempt to describe the metabolism of human cells focusing on breast cells. The model has two versions related to the presence or absence of a specific breast tumor. The aim will be to mine these genome-scale models as a multi-objective optimization problem in order to maximize biomass production and minimize the reactions whose enzymes that catalyze them may have undergone mutations (oncometabolite), causing cancer cells to proliferate. This study discovered characteristic pathological patterns for the breast cancer genome-scale model used. This work presents an in silico BioCAD methodology to investigate and compare the metabolic pathways of breast tissue in the presence of a tumor in contrast to those of healthy tissue. A large number of genome-scale metabolic model simulations have been carried out to explore the solution spaces of genetic configurations and metabolic reactions. An evolutionary algorithm is employed to guide the search for possible solutions, and a multi-objective optimization principle is used to identify the best candidate solutions.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Amaradio, M.N., Ojha, V., Jansen, G., Gulisano, M., Costanza, J., Nicosia, G.: Pareto optimal metabolic engineering for the growth-coupled overproduction of sustainable chemicals. Biotechnol. Bioeng. 119(7), 1890–1902 (2022)
Angione, C., Costanza, J., Carapezza, G., Lió, P., Nicosia, G.: A design automation framework for computational bioenergetics in biological networks. Mol. BioSyst. 9(10), 2554–2564 (2013)
Barzaman, K., et al.: Breast cancer: biology, biomarkers, and treatments. Int. Immunopharmacol. 84, 106535 (2020)
Biondi, T., Ciccazzo, A., Cutello, V., D’Antona, S., Nicosia, G., Spinella, S.: Multi-objective evolutionary algorithms and pattern search methods for circuit design problems. J. Univers. Comput. Sci. 12(4), 432–449 (2006)
Chou, F.J., Liu, Y., Lang, F., Yang, C.: D-2-hydroxyglutarate in glioma biology. Cells 10(9), 2345 (2021)
Cutello, V., Lee, D., Leone, S., Nicosia, G., Pavone, M.: Clonal selection algorithm with dynamic population size for bimodal search spaces. In: Jiao, L., Wang, L., Gao, X.-b, Liu, J., Wu, F. (eds.) ICNC 2006. LNCS, vol. 4221, pp. 949–958. Springer, Heidelberg (2006). https://doi.org/10.1007/11881070_125
Cutello, V., Nicosia, G., Pavone, M., Stracquadanio, G.: An information-theoretic approach for clonal selection algorithms. In: Hart, E., McEwan, C., Timmis, J., Hone, A. (eds.) ICARIS 2010. LNCS, vol. 6209, pp. 144–157. Springer, Heidelberg (2010). https://doi.org/10.1007/978-3-642-14547-6_12
DallaPozza, E., et al.: Regulation of succinate dehydrogenase and role of succinate in cancer. Semin. Cell Dev. Biol. 98, 4–14 (2020)
Ježek, P.: 2-Hydroxyglutarate in cancer cells. Antioxid. Redox Signal. 33(13), 903–926 (2020)
Katsura, C., Ogunmwonyi, I., Kankam, H.K., Saha, S.: Breast cancer: presentation, investigation, and management. Br. J. Hosp. Med. (London, England) 83(2), 1–7 (2005)
Keller, M.A., Piedrafita, G., Ralser, M.: The widespread role of non-enzymatic reactions in cellular metabolism. Curr. Opin. Biotechnol. 34, 153–161 (2015)
King, Z.A., et al.: BiGG models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44(D1), D515–D522 (2016)
Hucka, M., et al.: The systems biology markup language (SBML): language specification for level 3 version 2 Core release 2. J. Integr. Bioinform. 16(2), 20190021 (2019)
Liu, Y., et al.: Targeting tumor suppressor genes for cancer therapy. BioEssays: News Rev. Mol. Cell. Dev. Biol. 37(12), 1277–1286 (2015)
Liu, S., Cadoux-Hudson, T., Schofield, C.J.: Isocitrate dehydrogenase variants in cancer - cellular consequences and therapeutic opportunities. Curr. Opin. Chem. Biol. 57, 122–134 (2020)
Mishra, P., Ambs, S.: Metabolic signatures of human breast cancer. Mol. Cell. Oncol. 2(3), e992217 (2015)
Nam, H., et al.: A systems approach to predict oncometabolites via context-specific genome-scale metabolic networks. PLoS Comput. Biol. 10(9), e1003837 (2014)
Nicosia, G., Stracquadanio, G.: Generalized pattern search and mesh adaptive direct search algorithms for protein structure prediction. In: Giancarlo, R., Hannenhalli, S. (eds.) WABI 2007. LNCS, vol. 4645, pp. 183–193. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-74126-8_17
Norsigian, C.J., et al.: BiGG models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 48(D1), D402–D406 (2020)
Orth, J., Thiele, I., Palsson, B.: What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010)
Patanè, A., Santoro, A., Costanza, J., Carapezza, G., Nicosia, G.: Pareto optimal design for synthetic biology. IEEE Trans. Biomed. Circuits Syst. 9(4), 555–571 (2015)
Patané, A., Jansen, G., Conca, P., Carapezza, G., Costanza, J., Nicosia, G.: Multi-objective optimization of genome-scale metabolic models: the case of ethanol production. Ann. Oper. Res. 276(1–2), 211–227 (2018). https://doi.org/10.1007/s10479-018-2865-4
Peetsold, M., et al.: fumarase deficiency: a case with a new pathogenic mutation and a review of the literature. J. Child Neurol. 36(4), 310–323 (2021)
Rana, P., Berry, C., Ghosh, P., Fong, S.S.: Recent advances on constraint-based models by integrating machine learning. Curr. Opin. Biotechnol. 64, 85–91 (2020)
Sharifi, M.R., Akbarifard, S., Qaderi, K., Madadi, M.R.: A new optimization algorithm to solve multi-objective problems. Sci. Rep. 11(1), 20326 (2021)
Schmidt, C., Sciacovelli, M., Frezza, C.: Fumarate hydratase in cancer: a multifaceted tumor suppressor. Semin. Cell Dev. Biol. 98, 15–25 (2020)
Umeton, R., Nicosia, G., Dewey, C.F.: OREMPdb: a semantic dictionary of computational pathway models. BMC Bioinform. 13(4), 1–9 (2012)
Van Rosmalen, R.P., Smith, R.W., Martins Dos Santos, V.A.P., Fleck, C., Suarez-Diez, M.: Model reduction of genome-scale metabolic models as a basis for targeted kinetic models. Metab. Eng. 64, 74–84 (2021)
Vander Heiden, M.G.: Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 10(9), 671–684 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix
Appendix
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Amaradio, M.N., Jansen, G., Ojha, V., Costanza, J., Di Fatta, G., Nicosia, G. (2023). Inferring Pathological Metabolic Patterns in Breast Cancer Tissue from Genome-Scale Models. In: Nicosia, G., et al. Machine Learning, Optimization, and Data Science. LOD 2022. Lecture Notes in Computer Science, vol 13810. Springer, Cham. https://doi.org/10.1007/978-3-031-25599-1_43
Download citation
DOI: https://doi.org/10.1007/978-3-031-25599-1_43
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-25598-4
Online ISBN: 978-3-031-25599-1
eBook Packages: Computer ScienceComputer Science (R0)