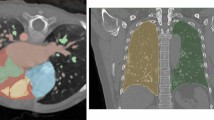
Abstract
In 3D medical image segmentation, small targets segmentation is crucial for diagnosis but still faces challenges. In this paper, we propose the Axis Projection Attention UNet, named APAUNet, for 3D medical image segmentation, especially for small targets. Considering the large proportion of the background in the 3D feature space, we introduce a projection strategy to project the 3D features into three orthogonal 2D planes to capture the contextual attention from different views. In this way, we can filter out the redundant feature information and mitigate the loss of critical information for small lesions in 3D scans. Then we utilize a dimension hybridization strategy to fuse the 3D features with attention from different axes and merge them by a weighted summation to adaptively learn the importance of different perspectives. Finally, in the APA Decoder, we concatenate both high and low resolution features in the 2D projection process, thereby obtaining more precise multi-scale information, which is vital for small lesion segmentation. Quantitative and qualitative experimental results on two public datasets (BTCV and MSD) demonstrate that our proposed APAUNet outperforms the other methods. Concretely, our APAUNet achieves an average dice score of 87.84 on BTCV, 84.48 on MSD-Liver and 69.13 on MSD-Pancreas, and significantly surpass the previous SOTA methods on small targets.
Y. Jiang and Z. Zhang—Equal contributions. Code is available at github.com/ zx33/APAUNet.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Zhu, Z., Xia, Y., Shen, W., Fishman, E., Yuille, A.: A 3D coarse-to-fine framework for volumetric medical image segmentation. In: 2018 International conference on 3D vision (3DV) (2018)
Wang, G., et al.: Automatic segmentation of vestibular schwannoma from T2-weighted MRI by deep spatial attention with hardness-weighted loss. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 264–272. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_30
Zheng, H., et al.: A new ensemble learning framework for 3D biomedical image segmentation. In: Proceedings of the Thirty-Third AAAI Conference on Artificial Intelligence and Thirty-First Innovative Applications of Artificial Intelligence Conference and Ninth AAAI Symposium on Educational Advances in Artificial Intelligence, pp. 5909–5916 (2019)
Xia, Y., Xie, L., Liu, F., Zhu, Z., Fishman, E.K., Yuille, A.L.: Bridging the gap between 2D and 3D organ segmentation with volumetric fusion net. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 445–453. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_51
Gao, Y., Zhou, M., Metaxas, D.N.: UTNet: a hybrid transformer architecture for medical image segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12903, pp. 61–71. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87199-4_6
Hatamizadeh, A., et al.: UneTR: transformers for 3D medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (2022)
Xie, Y., Zhang, J., Shen, C., Xia, Y.: CoTR: efficiently bridging CNN and transformer for 3D medical image segmentation. CoRR abs/2103.03024 (2021)
Landman, B., Xu, Z., Igelsias, J., Styner, M., Langerak, T., Klein, A.: MICCAI multi-atlas labeling beyond the cranial vault-workshop and challenge. In: Proceedings of MICCAI Multi-atlas Labeling Beyond Cranial Vault-Workshop Challenge, vol. 5, p. 12 (2015)
Simpson, A.L., Antonelli, M., Bakas, S., Bilello, M., Farahani, K., et al.: A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 (2019)
Xiao, X., Lian, S., Luo, Z., Li, S.: Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME) (2018)
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.W., Heng, P.A.: H-denseunet: hybrid densely connected unet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging (2018)
Yu, Q., et al.: C2fnas: coarse-to-fine neural architecture search for 3D medical image segmentation. In: Proceedings of CVPR (2020)
Jin, Q., Meng, Z., Sun, C., Cui, H., Su, R.: Ra-unet: a hybrid deep attention-aware network to extract liver and tumor in CT scans. Front. Bioeng. Biotechnol. (2020)
Oktay, O., Schlemper, J., Folgoc, L.L., Lee, M., Heinrich, M., et al.: Attention u-net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018)
Zhang, H., Cisse, M., Dauphin, Y.N., Lopez-Paz, D.: Mixup: beyond empirical risk minimization. In: International Conference on Learning Representations (2018)
Yun, S., Han, D., Oh, S.J., Chun, S., Choe, J., Yoo, Y.: Cutmix: regularization strategy to train strong classifiers with localizable features. In: Proceedings of ICCV (2019)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. (2017)
Xu, J., Li, M., Zhu, Z.: Automatic data augmentation for 3D medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (2020)
Huang, H., Lin, L., Tong, R., Hu, H., Zhang, Q., et al.: Unet 3+: a full-scale connected unet for medical image segmentation. In: Proceedings of ICASSP (2020)
He, Y., Yang, D., Roth, H., Zhao, C., Xu, D.: Dints: Differentiable neural network topology search for 3D medical image segmentation. In: Proceedings of CVPR. (2021)
Li, W., Qin, S., Li, F., Wang, L.: Mad-unet: a deep u-shaped network combined with an attention mechanism for pancreas segmentation in CT images. Med. Phys. (2021)
Li, Y., Yao, T., Pan, Y., Mei, T.: Contextual transformer networks for visual recognition. arXiv preprint arXiv:2107.12292 (2021)
Li, X., Wang, W., Hu, X., Yang, J.: Selective kernel networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 510–519 (2019)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Xie, E., et al.: Segmenting transparent object in the wild with transformer (2021)
Huang, X., Deng, Z., Li, D., Yuan, X.: Missformer: an effective medical image segmentation transformer. arXiv preprint arXiv:2109.07162 (2021)
Chen, J., et al.: Transunet: transformers make strong encoders for medical image segmentation. CoRR abs/2102.04306 (2021)
Isensee, F., et al.: NNU-net: self-adapting framework for u-net-based medical image segmentation. arXiv preprint arXiv:1809.10486 (2018)
Zhou, H.Y., Guo, J., Zhang, Y., Yu, L., Wang, L., Yu, Y.: NNFormer: interleaved transformer for volumetric segmentation. arXiv preprint arXiv:2109.03201 (2021)
Zheng, H., et al.: HFA-net: 3D cardiovascular image segmentation with asymmetrical pooling and content-aware fusion. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 759–767. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_84
Acknowledgements
This work was supported in part by the National Key R &D Program of China with grant No.2018YFB1800800, by the Basic Research Project No. HZQB-KCZYZ-2021067 of Hetao Shenzhen HK S &T Cooperation Zone, by NSFC-Youth 61902335, by Shenzhen Outstanding Talents Training Fund, by Guangdong Research Project No.2017ZT07X152 and No.2019CX01X104, by the Guangdong Provincial Key Laboratory of Future Networks of Intelligence (Grant No.2022B1212010001), by zelixir biotechnology company Fund, by the Guangdong Provincial Key Laboratory of Big Data Computing, The Chinese University of Hong Kong, Shenzhen, by Tencent Open Fund, and by ITSO at CUHKSZ.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jiang, Y., Zhang, Z., Qin, S., Guo, Y., Li, Z., Cui, S. (2023). APAUNet: Axis Projection Attention UNet for Small Target in 3D Medical Segmentation. In: Wang, L., Gall, J., Chin, TJ., Sato, I., Chellappa, R. (eds) Computer Vision – ACCV 2022. ACCV 2022. Lecture Notes in Computer Science, vol 13846. Springer, Cham. https://doi.org/10.1007/978-3-031-26351-4_2
Download citation
DOI: https://doi.org/10.1007/978-3-031-26351-4_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-26350-7
Online ISBN: 978-3-031-26351-4
eBook Packages: Computer ScienceComputer Science (R0)