Abstract
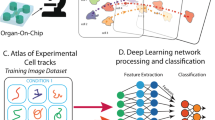
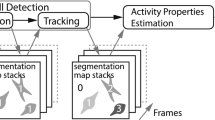
Cell motility, the ability of cells to move, is crucial in a wide range of biological processes; for instance, in cancer, it is directly related to metastasis. However, it is a complex phenomenon which is not well-understood yet, and studies are mainly done by human observation, which is subjective and error-prone. We intend to provide an automated mechanism to analyze the movement patterns that occur in in-vitro cell cultures, which can be registered by time lapse microscopy. Our approach, which is still a work in progress, utilizes an interactive 2D map that organizes motility patterns based on their similarity, enabling exploratory analysis. We extract the velocity fields that represent the cell displacements between consecutive frames and use a deep convolutional autoencoder to project a characterization of short video sequences of smaller parts of the original videos into a 10D latent space. The samples (small videos) are visualized in a 2D map using the Uniform Manifold Approximation and Projection (UMAP). The possibilities and extent of our method are showcased through a small interactive application that allows to explore all the types of cell motility patterns present in the training videos on a 2D map.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
The algorithm was implemented with Object Tracking of OpenCV-Python, using the following parameters: pyr_scale = 0.5, levels = 3, winsize = 60, iterations = 3, poly_n = 5, poly_sigma = 1.1, flags = cv2.OPTFLOW_FARNEBACK_GAUSSIAN.
- 2.
Head and Neck research group from the Instituto de Investigación Sanitaria del Principado de Asturias (ISPA, https://www.ispasturias.es).
References
Ali, M., Jones, M.W., Xie, X., Williams, M.: Timecluster: dimension reduction applied to temporal data for visual analytics. Vis. Comput. 35(6), 1013–1026 (2019)
Allaoui, M., Aissa, N.E.H.S.B., Belghith, A.B., Kherfi, M.L.: A machine learning-based tool for exploring covid-19 scientific literature. In: 2021 International Conference on Recent Advances in Mathematics and Informatics (ICRAMI), pp. 1–7. IEEE (2021)
Camley, B.A., Rappel, W.J.: Physical models of collective cell motility: from cell to tissue. J. Phys. D Appl. Phys. 50(11), 113002 (2017)
Chaudhry, R., Ravichandran, A., Hager, G., Vidal, R.: Histograms of oriented optical flow and binet-cauchy kernels on nonlinear dynamical systems for the recognition of human actions. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 1932–1939. IEEE (2009)
Colque, R.V.H.M., Caetano, C., de Andrade, M.T.L., Schwartz, W.R.: Histograms of optical flow orientation and magnitude and entropy to detect anomalous events in videos. IEEE Trans. Circuits Syst. Video Technol. 27(3), 673–682 (2016)
Cornejo, K.M., et al.: Succinate dehydrogenase B: a new prognostic biomarker in clear cell renal cell carcinoma. Hum. Pathol. 46(6), 820–826 (2015)
Fang, Z., Sun, Q., Yang, H., Zheng, J.: SDHB suppresses the tumorigenesis and development of ccRCC by inhibiting glycolysis. Front. Oncol. 11, 639408 (2021)
Farnebäck, G.: Two-frame motion estimation based on polynomial expansion. In: Bigun, J., Gustavsson, T. (eds.) SCIA 2003. LNCS, vol. 2749, pp. 363–370. Springer, Heidelberg (2003). https://doi.org/10.1007/3-540-45103-X_50
Hoshikawa, E., et al.: Cells/colony motion of oral keratinocytes determined by non-invasive and quantitative measurement using optical flow predicts epithelial regenerative capacity. Sci. Rep. 11(1), 1–12 (2021)
Huang, Y., Hao, L., Li, H., Liu, Z., Wang, P.: Quantitative analysis of intracellular motility based on optical flow model. J. Healthcare Eng. 2017, 1848314 (2017)
Kingma, D.P., Ba, J.: Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems 25 (2012)
Ladoux, B., Mège, R.M.: Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 18(12), 743–757 (2017)
Linehan, W.M., et al.: The metabolic basis of kidney cancer. Cancer Discov. 9(8), 1006–1021 (2019)
Liu, Y.J., Zhang, J.K., Yan, W.J., Wang, S.J., Zhao, G., Fu, X.: A main directional mean optical flow feature for spontaneous micro-expression recognition. IEEE Trans. Affect. Comput. 7(4), 299–310 (2015)
McConville, R., Santos-Rodriguez, R., Piechocki, R.J., Craddock, I.: N2d:(not too) deep clustering via clustering the local manifold of an autoencoded embedding. In: 2020 25th International Conference on Pattern Recognition (ICPR), pp. 5145–5152. IEEE (2021)
McInnes, L., Healy, J., Melville, J.: UMAP: uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426 (2018)
Morehead, A., Chantapakul, W., Cheng, J.: Semi-supervised graph learning meets dimensionality reduction. arXiv preprint arXiv:2203.12522 (2022)
Patterson, J., Gibson, A.: Deep learning: a practitioner’s approach. O’Reilly Media, Inc. (2017)
Ruder, S.: An overview of gradient descent optimization algorithms. arXiv preprint arXiv:1609.04747 (2016)
Stuelten, C.H., Parent, C.A., Montell, D.J.: Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat. Rev. Cancer 18(5), 296–312 (2018)
Te Boekhorst, V., Preziosi, L., Friedl, P.: Plasticity of cell migration in vivo and in silico. Annu. Rev. Cell Dev. Biol. 32(1), 491–526 (2016)
Thirion, J.P.: Image matching as a diffusion process: an analogy with Maxwell’s demons. Med. Image Anal. 2(3), 243–260 (1998). https://doi.org/10.1016/S1361-8415(98)80022-4
Verburg, M., Menkovski, V.: Micro-expression detection in long videos using optical flow and recurrent neural networks. In: 2019 14th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2019), pp. 1–6. IEEE (2019)
Vercauteren, T., Pennec, X., Perchant, A., Ayache, N.: Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage 45(1, Supplement 1), S61–S72 (2009). https://doi.org/10.1016/j.neuroimage.2008.10.040. https://www.sciencedirect.com/science/article/pii/S1053811908011683, mathematics in Brain Imaging
Vig, D.K., Hamby, A.E., Wolgemuth, C.W.: On the quantification of cellular velocity fields. Biophys. J . 110(7), 1469–1475 (2016)
Wang, Y., Yu, Z., Wang, Z.: A temporal clustering method fusing deep convolutional autoencoders and dimensionality reduction methods and its application in air quality visualization. Chemom. Intell. Lab. Syst. 227, 104607 (2022)
Yang, J., et al.: Functional deficiency of succinate dehydrogenase promotes tumorigenesis and development of clear cell renal cell carcinoma through weakening of ferroptosis. Bioengineered 13(4), 11187–11207 (2022)
Yong, C., Stewart, G.D., Frezza, C.: Oncometabolites in renal cancer. Nat. Rev. Nephrol. 16(3), 156–172 (2020)
Acknowledgments
This work was supported by the Ministerio de Ciencia e Innovación / Agencia Estatal de Investigación (MCIN/AEI/ 10.13039/501100011033) grant [PID2020-115401GB-I00]. The authors would also like to thank the financial support provided by the Principado de Asturias government through the predoctoral grant “Severo Ochoa”.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 IFIP International Federation for Information Processing
About this paper
Cite this paper
González, A. et al. (2023). Visualizing Cell Motility Patterns from Time Lapse Videos with Interactive 2D Maps Generated with Deep Autoencoders. In: Maglogiannis, I., Iliadis, L., Papaleonidas, A., Chochliouros, I. (eds) Artificial Intelligence Applications and Innovations. AIAI 2023 IFIP WG 12.5 International Workshops. AIAI 2023. IFIP Advances in Information and Communication Technology, vol 677. Springer, Cham. https://doi.org/10.1007/978-3-031-34171-7_37
Download citation
DOI: https://doi.org/10.1007/978-3-031-34171-7_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-34170-0
Online ISBN: 978-3-031-34171-7
eBook Packages: Computer ScienceComputer Science (R0)