Abstract
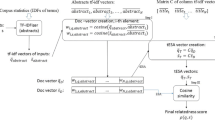
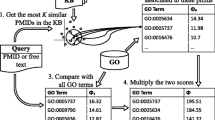
Gene ontology (GO) is a major source of biological knowledge that describes the functions of genes and gene products using a comprehensive set of controlled vocabularies or terms organized in a hierarchical structure. Automatic annotation of biological texts using gene ontology (GO) terms gained the attention of the scientific community as it helps to quickly identify relevant documents or parts of text related to specific biological functions or processes. In this paper, we propose and investigate a new GO-term annotation strategy that uses a non-parametric k-nearest neighbor model and relies on various vector-based representations of documents and GO terms linked to these documents. Our vector representations are based on machine learning and natural language processing (NLP) models, including singular value decomposition, Word2Vec and topic-based scoring. We evaluate the performance of our model on a large benchmark corpus using a variety of standard and hierarchical evaluation metrics.
Supported by the Defense Advanced Research Projects Agency (DARPA) through Cooperative Agreement D20AC00002 awarded by the U.S. Department of the Interior, Interior Business Center. The content of the article does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Arighi, C., et al.: Proceedings of the fourth biocreative challenge evaluation workshop (2013)
Blaschke, C., Leon, E.A., Krallinger, M., Valencia, A.: Evaluation of biocreative assessment of task 2. BMC Bioinform. 6, 1–13 (2005)
Camon, E.B., et al.: An evaluation of go annotation retrieval for biocreative and goa. BMC Bioinf. 6, 1–11 (2005)
Chen, Y.D., Yang, C.J., Li, W.G., Huang, C.Y., Chiang, J.H., et al.: Gene ontology evidence sentence extraction and concept extraction: two rule-based approaches (2013)
Faria, D., Schlicker, A., Pesquita, C., Bastos, H., Ferreira, A.E., Albrecht, M., Falcão, A.O.: Mining go annotations for improving annotation consistency. PLoS ONE 7(7), e40519 (2012)
Gobeill, J., Pasche, E., Vishnyakova, D., Ruch, P.: Closing the loop: from paper to protein annotation using supervised gene ontology classification. Database 2014 (2014)
Lena, P.D., Domeniconi, G., Margara, L., Moro, G.: Gota: Go term annotation of biomedical literature. BMC Bioinform. 16, 1–13 (2015)
Lu, Z., Hirschman, L.: Biocuration workflows and text mining: overview of the biocreative 2012 workshop track ii. Database 2012 (2012)
Voorhees, E.M., Buckland, L.: Overview of the trec 2003 question answering track. In: TREC, vol. 2003, pp. 54–68 (2003)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jui, J.H., Hauskrecht, M. (2023). Machine Learning Models for Automatic Gene Ontology Annotation of Biological Texts. In: Juarez, J.M., Marcos, M., Stiglic, G., Tucker, A. (eds) Artificial Intelligence in Medicine. AIME 2023. Lecture Notes in Computer Science(), vol 13897. Springer, Cham. https://doi.org/10.1007/978-3-031-34344-5_24
Download citation
DOI: https://doi.org/10.1007/978-3-031-34344-5_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-34343-8
Online ISBN: 978-3-031-34344-5
eBook Packages: Computer ScienceComputer Science (R0)