Abstract
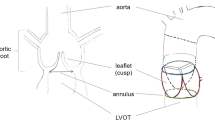
To perform aortic valve morphology for the assessment of the valvular heart disease in cardiovascular medicine, an accurate identification of specific anatomical points, i.e. landmarks, which define the aortic cusps, is required. In this study, we investigate the application of a deep learning framework, namely the spatial configuration network, for aortic cusp landmark detection in 120 contrast-enhanced end-diastolic coronary computed tomography images of normal patients. By performing three-fold cross-validation experiments, we obtained a mean detection error of \(1.45\,{\pm }\,0.82\) mm for six landmarks located at the nadirs and commissures of the aortic valve sinuses, which dropped to \(1.15\,{\pm }\,0.62\) mm when landmarks were detected in images that were cropped around the aortic valve by applying atlas-based segmentation. The obtained accuracy is comparable to existing methods, however, additional improvements in the form of image pre- or post-processing, or by applying advanced methodological concepts, may improve the landmark detection performance.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Al, W.A., Jung, H.Y., Yun, I.D., Jang, Y., Park, H.B., Chang, H.J.: Automatic aortic valve landmark localization in coronary CT angiography using colonial walk. PLoS ONE 13(7), e0200317 (2018). https://doi.org/10.1371/journal.pone.0200317
Aoyama, G., et al.: Automatic aortic valve cusps segmentation from CT images based on the cascading multiple deep neural networks. J. Imaging 8(1), 11 (2022). https://doi.org/10.3390/jimaging8010011
Bekkouch, I.E.I., Maksudov, B., Kiselev, S., Mustafaev, T., Vrtovec, T., Ibragimov, B.: Multi-landmark environment analysis with reinforcement learning for pelvic abnormality detection and quantification. Med. Image Anal. 78, 102417 (2022). https://doi.org/10.1016/j.media.2022.102417
Calleja, A., et al.: Automated quantitative 3-dimensional modeling of the aortic valve and root by 3-dimensional transesophageal echocardiography in normals, aortic regurgitation, and aortic stenosis. Circ. Cardiovasc. Imaging 6(1), 99–108 (2013). https://doi.org/10.1161/CIRCIMAGING.112.976993
Chen, C., et al.: Deep learning for cardiac image segmentation: a review. Front. Cardiovasc. Med. 7, 25 (2020). https://doi.org/10.3389/fcvm.2020.00025
Coffey, S., et al.: Global epidemiology of valvular heart disease. Nat. Rev. Cardiol. 18, 853–864 (2021). https://doi.org/10.1038/s41569-021-00570-z
Klein, S., Staring, M., Murphy, K., Viergever, M.A., Pluim, J.P.W.: elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29(1), 196–205 (2010). https://doi.org/10.1109/TMI.2009.2035616
Krittanawong, C., et al.: Deep learning for cardiovascular medicine: a practical primer. Eur. Heart J. 40(25), 2058–2073 (2019). https://doi.org/10.1093/eurheartj/ehz056
Noothout, J.M.H., et al.: Deep learning-based regression and classification for automatic landmark localization in medical images. IEEE Trans. Med. Imaging 39(12), 4011–4022 (2020). https://doi.org/10.1109/TMI.2020.3009002
Payer, C., Štern, D., Bischof, H., Urschler, M.: Integrating spatial configuration into heatmap regression based CNNs for landmark localization. Med. Image Anal. 54, 207–219 (2019). https://doi.org/10.1016/j.media.2019.03.007
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Tahoces, P.G., et al.: Automatic detection of anatomical landmarks of the aorta in CTA images. Med. Biol. Eng. Comput. 58(5), 903–919 (2020). https://doi.org/10.1007/s11517-019-02110-x
Tretter, J.T., et al.: Understanding the aortic root using computed tomographic assessment: a potential pathway to improved customized surgical repair. Circ. Cardiovasc. Imaging 14(11), e013134 (2021). https://doi.org/10.1161/CIRCIMAGING.121.013134
Yu, H., Yang, L.T., Zhang, Q., Armstrong, D., Deen, M.J.: Convolutional neural networks for medical image analysis: state-of-the art, comparisons, improvement and perspectives. Neurocomputing 444, 92–110 (2021). https://doi.org/10.1016/j.neucom.2020.04.157
Zhou, S.K., Le, H.N., Luu, K., Nguyen, H.V., Ayache, N.: Deep reinforcement learning in medical imaging: a literature review. Med. Image Anal. 73, 102193 (2021). https://doi.org/10.1016/j.media.2021.102193
Acknowledgements
The study was approved by the Ethics Committee of the University Medical Center Ljubljana, Slovenia, under 0120-133/2021/3 and 0120-312/2022/3, and supported by the Slovenian Research Agency (ARRS) under grants J2-4453 and P2-0232, and by the University Medical Center Ljubljana, Slovenia, under grant 20190174.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Škrlj, L., Jelenc, M., Vrtovec, T. (2023). Detection of Aortic Cusp Landmarks in Computed Tomography Images with Deep Learning. In: Bernard, O., Clarysse, P., Duchateau, N., Ohayon, J., Viallon, M. (eds) Functional Imaging and Modeling of the Heart. FIMH 2023. Lecture Notes in Computer Science, vol 13958. Springer, Cham. https://doi.org/10.1007/978-3-031-35302-4_31
Download citation
DOI: https://doi.org/10.1007/978-3-031-35302-4_31
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-35301-7
Online ISBN: 978-3-031-35302-4
eBook Packages: Computer ScienceComputer Science (R0)