Abstract
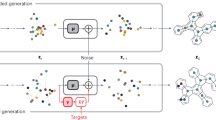
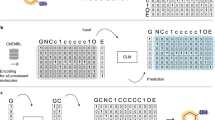
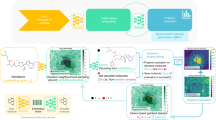
Optimizing molecular design and discovering novel chemical structures to meet specific objectives, such as quantitative estimates of the drug-likeness score (QEDs), is NP-hard due to the vast combinatorial design space of discrete molecular structures, which makes it near impossible to explore the entire search space comprehensively to exploit de novo structures with properties of interest. To address this challenge, reducing the intractable search space into a lower-dimensional latent volume helps examine molecular candidates more feasibly via inverse design. Autoencoders are suitable deep learning techniques, equipped with an encoder that reduces the discrete molecular structure into a latent space and a decoder that inverts the search space back to the molecular design. The continuous property of the latent space, which characterizes the discrete chemical structures, provides a flexible representation for inverse design to discover novel molecules. However, exploring this latent space requires particular insights to generate new structures. Therefore, we propose using a convex hull (CH) surrounding the top molecules regarding high QEDs to ensnare a tight subspace in the latent representation as an efficient way to reveal novel molecules with high QEDs. We demonstrate the effectiveness of our suggested method by using the QM9 as a training dataset along with the Self-Referencing Embedded Strings (SELFIES) representation to calibrate the autoencoder in order to carry out the inverse molecular design that leads to unfolding novel chemical structure.
This project is supported by the National Research Council Canada (NRC) and the Defence Research and Development Canada (DRDC).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abadi, M., et al.: \(\{\)TensorFlow\(\}\): a system for \(\{\)Large-Scale\(\}\) machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 2016), pp. 265–283 (2016)
Becke, A.D.: Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98(7), 5648–5652 (1993)
Blaschke, T., Olivecrona, M., Engkvist, O., Bajorath, J., Chen, H.: Application of generative autoencoder in de novo molecular design. Mol. Inf. 37(1–2), 1700123 (2018)
Ditchfield, R., Hehre, W.J., Pople, J.A.: Self-consistent molecular-orbital methods. IX. An extended gaussian-type basis for molecular-orbital studies of organic molecules. J. Chem. Phys. 54(2), 724–728 (1971)
Frisch, M.J., et al.: Gaussian 16 Revision C.01. Gaussian Inc., Wallingford (2016)
Ghaemi, M.S., Grantham, K., Tamblyn, I., Li, Y., Ooi, H.K.: Generative enriched sequential learning (ESL) approach for molecular design via augmented domain knowledge. In: Proceedings of the Canadian Conference on Artificial Intelligence, 27 May 2022
Grantham, K., Mukaidaisi, M., Ooi, H.K., Ghaemi, M.S., Tchagang, A., Li, Y.: Deep evolutionary learning for molecular design. IEEE Comput. Intell. Mag. 17(2), 14–28 (2022)
Joswig, M., Kaluba, M., Ruff, L.: Geometric disentanglement by random convex polytopes. arXiv preprint arXiv:2009.13987 (2020)
Kingma, D., Welling, M.: Auto-encoding variational Bayes. In: International Conference on Learning Representations (2014)
Lee, C., Yang, W., Parr, R.G.: Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785–789 (1988)
Menon, D., Ranganathan, R.: A generative approach to materials discovery, design, and optimization. ACS Omega 7(30), 25958–25973 (2022)
Ramakrishnan, R., Dral, P.O., Rupp, M., von Lilienfeld, O.A.: Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 1(1), 140022 (2014)
Romez-Bombarelli, R., et al.: Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018)
Sanchez-Lengeling, B., Aspuru-Guzik, A.: Inverse molecular design using machine learning: generative models for matter engineering. Science 361(6400), 360–365 (2018)
Vershynin, R.: High-Dimensional Probability. University of California, Irvine (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Ghaemi, M.S., Hu, H., Hu, A., Ooi, H.K. (2023). \(\textbf{CHA}_2\): CHemistry Aware Convex Hull Autoencoder Towards Inverse Molecular Design. In: Seipel, D., Steen, A. (eds) KI 2023: Advances in Artificial Intelligence. KI 2023. Lecture Notes in Computer Science(), vol 14236. Springer, Cham. https://doi.org/10.1007/978-3-031-42608-7_3
Download citation
DOI: https://doi.org/10.1007/978-3-031-42608-7_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-42607-0
Online ISBN: 978-3-031-42608-7
eBook Packages: Computer ScienceComputer Science (R0)