Abstract
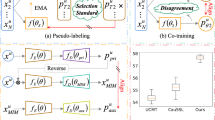
Medical image analysis using deep learning is often challenged by limited labeled data and high annotation costs. Fine-tuning the entire network in label-limited scenarios can lead to overfitting and suboptimal performance. Recently, prompt tuning has emerged as a more promising technique that introduces a few additional tunable parameters as prompts to a task-agnostic pre-trained model, and updates only these parameters using supervision from limited labeled data while keeping the pre-trained model unchanged. However, previous work has overlooked the importance of selective labeling in downstream tasks, which aims to select the most valuable downstream samples for annotation to achieve the best performance with minimum annotation cost. To address this, we propose a framework that combines selective labeling with prompt tuning (SLPT) to boost performance in limited labels. Specifically, we introduce a feature-aware prompt updater to guide prompt tuning and a TandEm Selective LAbeling (TESLA) strategy. TESLA includes unsupervised diversity selection and supervised selection using prompt-based uncertainty. In addition, we propose a diversified visual prompt tuning strategy to provide multi-prompt-based discrepant predictions for TESLA. We evaluate our method on liver tumor segmentation and achieve state-of-the-art performance, outperforming traditional fine-tuning with only 6% of tunable parameters, also achieving 94% of full-data performance by labeling only 5% of the data.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bilic, P., et al.: The liver tumor segmentation benchmark (LiTS). Med. Image Anal. 84, 102680 (2023)
Allingham, J.U., et al.: A simple zero-shot prompt weighting technique to improve prompt ensembling in text-image models. arXiv preprint arXiv:2302.06235 (2023)
Bai, F., Xing, X., Shen, Y., Ma, H., Meng, M.Q.H.: Discrepancy-based active learning for weakly supervised bleeding segmentation in wireless capsule endoscopy images. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds.) MICCAI 2022. LNCS, vol. 13438, pp. 24–34. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-16452-1_3
Beluch, W.H., Genewein, T., Nürnberger, A., Köhler, J.M.: The power of ensembles for active learning in image classification. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 9368–9377 (2018)
Brown, T.B., et al.: Language models are few-shot learners. In: Advances in Neural Information Processing Systems, pp. 1876–1901 (2020)
Caramalau, R., Bhattarai, B., Kim, T.K.: Sequential graph convolutional network for active learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9583–9592 (2021)
Chen, L.C., Papandreou, G., Schroff, F., Adam, H.: Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587 (2017)
Cheplygina, V., de Bruijne, M., Pluim, J.P.: Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 54, 280–296 (2019)
Dai, C., et al.: Suggestive annotation of brain tumour images with gradient-guided sampling. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12264, pp. 156–165. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59719-1_16
Gal, Y., Ghahramani, Z.: Dropout as a Bayesian approximation: representing model uncertainty in deep learning. In: International Conference on Machine Learning, pp. 1050–1059. PMLR (2016)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141 (2018)
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021)
Jia, M., et al.: Visual prompt tuning. In: Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T. (eds.) ECCV 2022. LNCS, vol. 13693, pp. 709–727. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-19827-4_41
Kim, M., et al.: Deep learning in medical imaging. Neurospine 16(4), 657 (2019)
Kullback, S., Leibler, R.A.: On information and sufficiency. Ann. Math. Stat. 22(1), 79–86 (1951)
Kumar, A., Raghunathan, A., Jones, R., Ma, T., Liang, P.: Fine-tuning can distort pretrained features and underperform out-of-distribution. arXiv preprint arXiv:2202.10054 (2022)
Liu, L., Yu, B.X., Chang, J., Tian, Q., Chen, C.W.: Prompt-matched semantic segmentation. arXiv preprint arXiv:2208.10159 (2022)
Liu, P., Yuan, W., Fu, J., Jiang, Z., Hayashi, H., Neubig, G.: Pre-train, prompt, and predict: a systematic survey of prompting methods in natural language processing. ACM Comput. Surv. 55(9), 1–35 (2023)
Parvaneh, A., Abbasnejad, E., Teney, D., Haffari, G.R., Van Den Hengel, A., Shi, J.Q.: Active learning by feature mixing. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12237–12246 (2022)
Powers, D.M.: Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061 (2020)
Salehi, S.S.M., Erdogmus, D., Gholipour, A.: Tversky loss function for image segmentation using 3D fully convolutional deep networks. In: Wang, Q., Shi, Y., Suk, H.-I., Suzuki, K. (eds.) MLMI 2017. LNCS, vol. 10541, pp. 379–387. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67389-9_44
Sener, O., Savarese, S.: Active learning for convolutional neural networks: a core-set approach. arXiv preprint arXiv:1708.00489 (2017)
Settles, B.: Active learning literature survey (2009)
Simpson, A.L., et al.: A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 (2019)
Tajbakhsh, N., Jeyaseelan, L., Li, Q., Chiang, J.N., Wu, Z., Ding, X.: Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med. Image Anal. 63, 101693 (2020)
Wang, T., et al.: Boosting active learning via improving test performance. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 36, pp. 8566–8574 (2022)
Zhan, X., Wang, Q., Huang, K.H., Xiong, H., Dou, D., Chan, A.B.: A comparative survey of deep active learning. arXiv preprint arXiv:2203.13450 (2022)
Zhao, T., et al.: Prompt design for text classification with transformer-based models. In: Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, pp. 2709–2722 (2021)
Zhou, K., Yang, J., Loy, C.C., Liu, Z.: Learning to prompt for vision-language models. Int. J. Comput. Vision 130(9), 2337–2348 (2022)
Acknowledgements
The work was supported by Alibaba Research Intern Program. Fan Bai and Max Q.-H. Meng were supported by National Key R &D program of China with Grant No. 2019YFB1312400, Hong Kong RGC CRF grant C4063-18G, and Hong Kong Health and Medical Research Fund (HMRF) under Grant 06171066. Xiaoli Yin and Yu Shi were supported by National Natural Science Foundation of China (82071885).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Bai, F. et al. (2023). SLPT: Selective Labeling Meets Prompt Tuning on Label-Limited Lesion Segmentation. In: Greenspan, H., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14221. Springer, Cham. https://doi.org/10.1007/978-3-031-43895-0_2
Download citation
DOI: https://doi.org/10.1007/978-3-031-43895-0_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43894-3
Online ISBN: 978-3-031-43895-0
eBook Packages: Computer ScienceComputer Science (R0)