Abstract
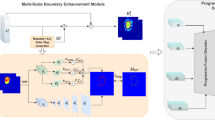
Skin lesion segmentation in dermoscopy images has seen recent success due to advancements in multi-scale boundary attention and feature-enhanced modules. However, existing methods that rely on end-to-end learning paradigms, which directly input images and output segmentation maps, often struggle with extremely hard boundaries, such as those found in lesions of particularly small or large sizes. This limitation arises because the receptive field and local context extraction capabilities of any finite model are inevitably limited, and the acquisition of additional expert-labeled data required for larger models is costly. Motivated by the impressive advances of diffusion models that regard image synthesis as a parameterized chain process, we introduce a novel approach that formulates skin lesion segmentation as a boundary evolution process to thoroughly investigate the boundary knowledge. Specifically, we propose the Medical Boundary Diffusion Model (MB-Diff), which starts with a randomly sampled Gaussian noise, and the boundary evolves within finite times to obtain a clear segmentation map. First, we propose an efficient multi-scale image guidance module to constrain the boundary evolution, which makes the evolution direction suit our desired lesions. Second, we propose an evolution uncertainty-based fusion strategy to refine the evolution results and yield more precise lesion boundaries. We evaluate the performance of our model on two popular skin lesion segmentation datasets and compare our model to the latest CNN and transformer models. Our results demonstrate that our model outperforms existing methods in all metrics and achieves superior performance on extremely challenging skin lesions. The proposed approach has the potential to significantly enhance the accuracy and reliability of skin lesion segmentation, providing critical information for diagnosis and treatment. All resources will be publicly available at https://github.com/jcwang123/MBDiff.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Azad, R., Asadi-Aghbolaghi, M., Fathy, M., Escalera, S.: Attention Deeplabv3+: multi-level context attention mechanism for skin lesion segmentation. In: Bartoli, A., Fusiello, A. (eds.) ECCV 2020. LNCS, vol. 12535, pp. 251–266. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-66415-2_16
Cao, W., et al.: ICL-Net: global and local inter-pixel correlations learning network for skin lesion segmentation. IEEE J. Biomed. Health Inf. 27(1), 145–156 (2022)
Chen, J., et al.: TransUNet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
Czolbe, S., Arnavaz, K., Krause, O., Feragen, A.: Is segmentation uncertainty useful? In: Feragen, A., Sommer, S., Schnabel, J., Nielsen, M. (eds.) IPMI 2021. LNCS, vol. 12729, pp. 715–726. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-78191-0_55
DeVries, T., Taylor, G.W.: Leveraging uncertainty estimates for predicting segmentation quality. arXiv preprint arXiv:1807.00502 (2018)
Dhariwal, P., Nichol, A.: Diffusion models beat GANs on image synthesis. Adv. Neural Inf. Process. Syst. 34, 8780–8794 (2021)
Gu, R., et al.: Ca-net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans. Med. Imaging 40(2), 699–711 (2020)
Gutman, D., et al.: Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv preprint arXiv:1605.01397 (2016)
Ho, J., Jain, A., Abbeel, P.: Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 33, 6840–6851 (2020)
Li, H., et al.: Dense deconvolutional network for skin lesion segmentation. IEEE J. Biomed. Health Inf. 23(2), 527–537 (2018)
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2117–2125 (2017)
Mehrtash, A., Wells, W.M., Tempany, C.M., Abolmaesumi, P., Kapur, T.: Confidence calibration and predictive uncertainty estimation for deep medical image segmentation. IEEE Trans. Med. Imaging 39(12), 3868–3878 (2020)
Mendonça, T., Ferreira, P.M., Marques, J.S., Marcal, A.R., Rozeira, J.: PH 2-A dermoscopic image database for research and benchmarking. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 5437–5440. IEEE (2013)
Nichol, A.Q., Dhariwal, P.: Improved denoising diffusion probabilistic models. In: International Conference on Machine Learning, pp. 8162–8171. PMLR (2021)
Rombach, R., Blattmann, A., Lorenz, D., Esser, P., Ommer, B.: High-resolution image synthesis with latent diffusion models. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 10684–10695 (2022)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Siegel, R.L., Miller, K.D., Fuchs, H.E., Jemal, A.: Cancer statistics, 2022. CA: Cancer J. Clin. 72(1), 7–33 (2022)
Wang, J., et al.: XBound-former: toward cross-scale boundary modeling in transformers. IEEE Trans. Med. Imaging 42(6), 1735–1745 (2023)
Wang, J., Wei, L., Wang, L., Zhou, Q., Zhu, L., Qin, J.: Boundary-aware transformers for skin lesion segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 206–216. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_20
Wang, W., et al.: Pyramid vision transformer: a versatile backbone for dense prediction without convolutions. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 568–578 (2021)
Wu, H., Pan, J., Li, Z., Wen, Z., Qin, J.: Automated skin lesion segmentation via an adaptive dual attention module. IEEE Trans. Med. Imaging 40(1), 357–370 (2020)
Wu, J., Fang, H., Zhang, Y., Yang, Y., Xu, Y.: MedSegDiff: Medical image segmentation with diffusion probabilistic model. arXiv preprint arXiv:2211.00611 (2022)
Zhang, Y., Liu, H., Hu, Q.: TransFuse: fusing transformers and CNNs for medical image segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 14–24. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_2
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: UNet++: a nested u-net architecture for medical image segmentation. In: Stoyanov, D., et al. (eds.) DLMIA/ML-CDS -2018. LNCS, vol. 11045, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00889-5_1
Acknowledgement
This work is supported by the National Key Research and Development Program of China (2019YFE0113900).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, J., Yang, J., Zhou, Q., Wang, L. (2023). Medical Boundary Diffusion Model for Skin Lesion Segmentation. In: Greenspan, H., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14223. Springer, Cham. https://doi.org/10.1007/978-3-031-43901-8_41
Download citation
DOI: https://doi.org/10.1007/978-3-031-43901-8_41
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43900-1
Online ISBN: 978-3-031-43901-8
eBook Packages: Computer ScienceComputer Science (R0)