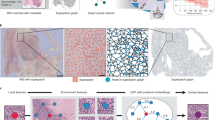
Abstract
In computation pathology, the pyramid structure of gigapixel Whole Slide Images (WSIs) has recently been studied for capturing various information from individual cell interactions to tissue microenvironments. This hierarchical structure is believed to be beneficial for cancer diagnosis and prognosis tasks. However, most previous hierarchical WSI analysis works (1) only characterize local or global correlations within the WSI pyramids and (2) use only unidirectional interaction between different resolutions, leading to an incomplete picture of WSI pyramids. To this end, this paper presents a novel Hierarchical Interaction Graph-Transformer (i.e., HIGT) for WSI analysis. With Graph Neural Network and Transformer as the building commons, HIGT can learn both short-range local information and long-range global representation of the WSI pyramids. Considering that the information from different resolutions is complementary and can benefit each other during the learning process, we further design a novel Bidirectional Interaction block to establish communication between different levels within the WSI pyramids. Finally, we aggregate both coarse-grained and fine-grained features learned from different levels together for slide-level prediction. We evaluate our methods on two public WSI datasets from TCGA projects, i.e., kidney carcinoma (KICA) and esophageal carcinoma (ESCA). Experimental results show that our HIGT outperforms both hierarchical and non-hierarchical state-of-the-art methods on both tumor subtyping and staging tasks.
Z. Guo and W. Zhao: Contributed equally to this work.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
AbdulJabbar, K., et al.: Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nature Med. 26(7), 1054–1062 (2020)
Brody, S., Alon, U., Yahav, E.: How attentive are graph attention networks? arXiv preprint arXiv:2105.14491 (2021)
Chen, R.J., et al.: Scaling vision transformers to gigapixel images via hierarchical self-supervised learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 16144–16155 (June 2022)
Chen, R.J., et al.: Whole slide images are 2D Point Clouds: context-aware survival prediction using patch-based graph convolutional networks. In: de Bruijne, M., et al. (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2021: 24th International Conference, Strasbourg, France, September 27 – October 1, 2021, Proceedings, Part VIII, pp. 339–349. Springer International Publishing, Cham (2021). https://doi.org/10.1007/978-3-030-87237-3_33
Fey, M., Lenssen, J.E.: Fast graph representation learning with pytorch geometric. arXiv preprint arXiv:1903.02428 (2019)
Hou, W., Wang, L., Cai, S., Lin, Z., Yu, R., Qin, J.: Early neoplasia identification in Barrett’s esophagus via attentive hierarchical aggregation and self-distillation. Medical Image Anal. 72, 102092 (2021). https://doi.org/10.1016/j.media.2021.102092
Hou, W., et al.: \(\text{H}^2\)-mil: Exploring hierarchical representation with heterogeneous multiple instance learning for whole slide image analysis. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 36, pp. 933–941 (2022)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141 (2018)
Ilse, M., Tomczak, J., Welling, M.: Attention-based deep multiple instance learning. In: International Conference on Machine Learning, pp. 2127–2136. PMLR (2018)
Kumar, N., Verma, R., Sharma, S., Bhargava, S., Vahadane, A., Sethi, A.: A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 36(7), 1550–1560 (2017)
Li, B., Li, Y., Eliceiri, K.W.: Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 14318–14328 (2021)
Lu, M.Y., Williamson, D.F., Chen, T.Y., Chen, R.J., Barbieri, M., Mahmood, F.: Data-efficient and weakly supervised computational pathology on whole-slide images. Nature Biomed. Eng. 5(6), 555–570 (2021)
Mehta, S., Rastegari, M.: Separable self-attention for mobile vision transformers. arXiv preprint arXiv:2206.02680 (2022)
Paszke, A., et al.: Pytorch: an imperative style, high-performance deep learning library. Adv. Neural Inform. Process. Syst. 32 (2019)
Reisenbüchler, D., Wagner, S.J., Boxberg, M., Peng, T.: Local attention graph-based transformer for multi-target genetic alteration prediction. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022: 25th International Conference, Singapore, September 18–22, 2022, Proceedings, Part II, pp. 377–386. Springer Nature Switzerland, Cham (2022). https://doi.org/10.1007/978-3-031-16434-7_37
Riasatian, A., et al.: Fine-tuning and training of densenet for histopathology image representation using TCGA diagnostic slides. Med. Image Anal. 70, 102032 (2021)
Wu, Z., Jain, P., Wright, M., Mirhoseini, A., Gonzalez, J.E., Stoica, I.: Representing long-range context for graph neural networks with global attention. Adv. Neural. Inf. Process. Syst. 34, 13266–13279 (2021)
Yao, J., Zhu, X., Jonnagaddala, J., Hawkins, N., Huang, J.: Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med. Image Anal. 65, 101789 (2020)
Yao, X.H., et al.: Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 30(6), 541–543 (2020)
Zheng, Y., et al.: A graph-transformer for whole slide image classification. IEEE Trans. Med. Imaging 41(11), 3003–3015 (2022)
Acknowledgement
The work described in this paper was partially supported by grants from the National Natural Science Fund (62201483), the Research Grants Council of the Hong Kong Special Administrative Region, China (T45-401/22-N), and The Hong Kong Polytechnic University (P0045999).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Guo, Z., Zhao, W., Wang, S., Yu, L. (2023). HIGT: Hierarchical Interaction Graph-Transformer for Whole Slide Image Analysis. In: Greenspan, H., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14225. Springer, Cham. https://doi.org/10.1007/978-3-031-43987-2_73
Download citation
DOI: https://doi.org/10.1007/978-3-031-43987-2_73
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43986-5
Online ISBN: 978-3-031-43987-2
eBook Packages: Computer ScienceComputer Science (R0)