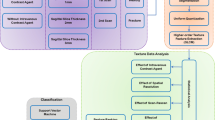
Abstract
Sarcopenia is a condition of age-associated muscle degeneration that shortens the life expectancy in those it affects, compared to individuals with normal muscle strength. Accurate screening for sarcopenia is a key process of clinical diagnosis and therapy. In this work, we propose a novel multi-modality contrastive learning (MM-CL) based method that combines hip X-ray images and clinical parameters for sarcopenia screening. Our method captures the long-range information with Non-local CAM Enhancement, explores the correlations in visual-text features via Visual-text Feature Fusion, and improves the model’s feature representation ability through Auxiliary contrastive representation. Furthermore, we establish a large in-house dataset with 1,176 patients to validate the effectiveness of multi-modality based methods. Significant performances with an AUC of 84.64%, ACC of 79.93%, F1 of 74.88%, SEN of 72.06%, SPC of 86.06%, and PRE of 78.44%, show that our method outperforms other single-modality and multi-modality based methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Source code will be released at https://github.com/qgking/MM-CL.git.
References
Ackermans, L.L., et al.: Screening, diagnosis and monitoring of sarcopenia: when to use which tool? Clinical Nutrition ESPEN (2022)
Braman, N., Gordon, J.W.H., Goossens, E.T., Willis, C., Stumpe, M.C., Venkataraman, J.: Deep orthogonal fusion: multimodal prognostic biomarker discovery integrating radiology, pathology, genomic, and clinical data. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12905, pp. 667–677. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87240-3_64
Chen, R.J., et al.: Pathomic fusion: an integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans. Med. Imaging 41(4), 757–770 (2020)
Chen, T., Kornblith, S., Norouzi, M., Hinton, G.: A simple framework for contrastive learning of visual representations. In: International Conference on Machine Learning, pp. 1597–1607. PMLR (2020)
Cruz-Jentoft, A.J., et al.: Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 48(1), 16–31 (2019)
Cubuk, E.D., Zoph, B., Mane, D., Vasudevan, V., Le, Q.V.: AutoAugment: learning augmentation strategies from data. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 113–123 (2019)
Dodds, R.M., Granic, A., Davies, K., Kirkwood, T.B., Jagger, C., Sayer, A.A.: Prevalence and incidence of sarcopenia in the very old: findings from the Newcastle 85+ study. J. Cachexia, Sarcopenia Muscle 8(2), 229–237 (2017)
Giovannini, S., et al.: Sarcopenia: diagnosis and management, state of the art and contribution of ultrasound. J. Clin. Med. 10(23), 5552 (2021)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Omeiza, D., Speakman, S., Cintas, C., Weldermariam, K.: Smooth grad-CAM++: an enhanced inference level visualization technique for deep convolutional neural network models. arXiv preprint arXiv:1908.01224 (2019)
Pang, B.W.J., et al.: Prevalence and associated factors of Sarcopenia in Singaporean adults-the Yishun Study. J. Am. Med. Direct. Assoc. 22(4), e1-885 (2021)
Ryu, J., Eom, S., Kim, H.C., Kim, C.O., Rhee, Y., You, S.C., Hong, N.: Chest X-ray-based opportunistic screening of sarcopenia using deep learning. J. Cachexia, Sarcopenia Muscle 14(1), 418–428 (2022)
Shafiee, G., Keshtkar, A., Soltani, A., Ahadi, Z., Larijani, B., Heshmat, R.: Prevalence of sarcopenia in the world: a systematic review and meta-analysis of general population studies. J. Diab. Metab. Disord. 16(1), 1–10 (2017)
Van Der Maaten, L.: Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15(1), 3221–3245 (2014)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, vol. 30 (2017)
Wang, X., Girshick, R., Gupta, A., He, K.: Non-local neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7794–7803 (2018)
Yan, K., Guo, Y., Liu, B.: Pretp-2l: identification of therapeutic peptides and their types using two-layer ensemble learning framework. Bioinformatics 39(4), btad125 (2023)
Yan, K., Lv, H., Guo, Y., Peng, W., Liu, B.: samppred-gat: prediction of antimicrobial peptide by graph attention network and predicted peptide structure. Bioinformatics 39(1), btac715 (2023)
Zhang, J., Xie, Y., Xia, Y., Shen, C.: Attention residual learning for skin lesion classification. IEEE Trans. Med. Imaging 38(9), 2092–2103 (2019)
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2921–2929 (2016)
Acknowledgment
This work was supported by the Fundamental Research Funds for the Central Universities, the National Natural Science Foundation of China [Grant No. 62201460 and No. 62072329], and the National Key Technology R &D Program of China [Grant No. 2018YFB1701700].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jin, Q. et al. (2023). Multi-modality Contrastive Learning for Sarcopenia Screening from Hip X-rays and Clinical Information. In: Greenspan, H., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14225. Springer, Cham. https://doi.org/10.1007/978-3-031-43987-2_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-43987-2_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43986-5
Online ISBN: 978-3-031-43987-2
eBook Packages: Computer ScienceComputer Science (R0)