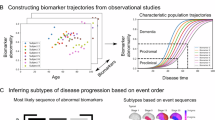
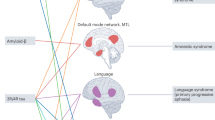
Abstract
Computational models of neurodegeneration aim to emulate the evolving pattern of pathology in the brain during neurodegenerative disease, such as Alzheimer’s disease. Previous studies have made specific choices on the mechanisms of pathology production and diffusion, or assume that all the subjects lie on the same disease progression trajectory. However, the complexity and heterogeneity of neurodegenerative pathology suggests that multiple mechanisms may contribute synergistically with complex interactions, meanwhile the degree of contribution of each mechanism may vary among individuals. We thus put forward a coupled-mechanisms modelling framework which non-linearly combines the network-topology-informed pathology appearance with the process of pathology spreading within a dynamic modelling system. We account for the heterogeneity of disease by fitting the model at the individual level, allowing the epicenters and rate of progression to vary among subjects. We construct a Bayesian model selection framework to account for feature importance and parameter uncertainty. This provides a combination of mechanisms that best explains the observations for each individual. With the obtained distribution of mechanism importance for each subject, we are able to identify subgroups of patients sharing similar combinations of apparent mechanisms.
E. Thompson and A. Schroder—Contributed equally to this work as the co-second authors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
References
Appel, S.H.: A unifying hypothesis for the cause of amyotrophic lateral sclerosis, parkinsonism, and Alzheimer disease. Ann. Neurol. J. Am. Neurol. Assoc. Child Neurol. Soc. 10(6), 499–505 (1981)
Bingham, E., et al.: Pyro: deep universal probabilistic programming. J. Mach. Learn. Res. 20(1), 973–978 (2019)
Buckner, R.L., et al.: Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J. Neurosci. 25(34), 7709 (2005). https://doi.org/10.1523/JNEUROSCI.2177-05.2005
Carvalho, C.M., Polson, N.G., Scott, J.G.: Handling sparsity via the horseshoe. In: Artificial Intelligence and Statistics, pp. 73–80. PMLR (2009)
Desikan, R.S., et al.: An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31(3), 968–980 (2006).https://doi.org/10.1016/J.NEUROIMAGE.2006.01.021
Garbarino, S., Lorenzi, M.: Investigating hypotheses of neurodegeneration by learning dynamical systems of protein propagation in the brain. NeuroImage 235, 117980 (2021). https://doi.org/10.1016/j.neuroimage.2021.117980
Garbarino, S., et al.: Differences in topological progression profile among neurodegenerative diseases from imaging data. eLife 8, e49298 (2019). https://doi.org/10.7554/eLife.49298
Groot, C., Villeneuve, S., Smith, R., Hansson, O., Ossenkoppele, R.: Tau PET imaging in neurodegenerative disorders. J. Nucl. Med. 63(Supplement 1), 20S-26S (2022). https://doi.org/10.2967/jnumed.121.263196
Iturria-Medina, Y., Carbonell, F.M., Evans, A.C., Initiative, A.D.N.: Multimodal imaging-based therapeutic fingerprints for optimizing personalized interventions: application to neurodegeneration. Neuroimage 179, 40–50 (2018)
Iturria-Medina, Y., Carbonell, F.M., Sotero, R.C., Chouinard-Decorte, F., Evans, A.C., Initiative, A.D.N.: Multifactorial causal model of brain (DIS) organization and therapeutic intervention: application to Alzheimer’s disease. Neuroimage 152, 60–77 (2017)
Iturria-Medina, Y., Sotero, R.C., Toussaint, P.J., Evans, A.C., Initiative, A.D.N.: Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders. PLoS Comput. Biol. 10(11), e1003956 (2014)
Jack, C.R., Jr., et al.: Predicting future rates of tau accumulation on PET. Brain 143(10), 3136–3150 (2020)
Landau, S., Jagust, W.: Flortaucipir (AV-1451) processing methods. Alzheimer’s Disease Neuroimaging Initiative (2016)
Meisl, G., et al.: In vivo rate-determining steps of tau seed accumulation in Alzheimer’s disease. Sci. Adv. 7, eabh1448 (2021)
Raj, A., Kuceyeski, A., Weiner, M.: A Network diffusion model of disease progression in dementia. Neuron 73(6), 1204–1215 (2012). https://doi.org/10.1016/j.neuron.2011.12.040
Raj, A., LoCastro, E., Kuceyeski, A., Tosun, D., Relkin, N., Weiner, M.: Network diffusion model of progression predicts longitudinal patterns of atrophy and metabolism in Alzheimer’s disease. Cell Rep. 10(3), 359–369 (2015)
Royer, J., et al.: An open MRI dataset for multiscale neuroscience. Sci. Data 9(1), 569 (2022). https://doi.org/10.1038/s41597-022-01682-y
Rubinov, M., Sporns, O.: Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52(3), 1059–1069 (2010)
Smith, R.E., Tournier, J.D., Calamante, F., Connelly, A.: SIFT2: enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. Neuroimage 119, 338–351 (2015). https://doi.org/10.1016/j.neuroimage.2015.06.092
Smith, R., et al.: The accumulation rate of tau aggregates is higher in females and younger amyloid-positive subjects. Brain 143(12), 3805–3815 (2020)
Tournier, J.D., et al.: MRtrix3: a fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 202, 116137 (2019). https://doi.org/10.1016/j.neuroimage.2019.116137
Van Essen, D.C., Smith, S.M., Barch, D.M., Behrens, T.E.J., Yacoub, E., Ugurbil, K.: WU-Minn HCP consortium: the WU-Minn human connectome project: an overview. Neuroimage 80, 62–79 (2013). https://doi.org/10.1016/j.neuroimage.2013.05.041
Vogel, J.W., et al.: Spread of pathological tau proteins through communicating neurons in human Alzheimer’s disease. Nat. Commun. 11(1), 2612 (2020). https://doi.org/10.1038/s41467-020-15701-2
Vogel, J.W., et al.: Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nat. Med. 27(5), 871–881 (2021). https://doi.org/10.1038/s41591-021-01309-6
Weickenmeier, J., Kuhl, E., Goriely, A.: Multiphysics of Prionlike diseases: progression and atrophy. Phys. Rev. Lett. 121(15), 158101 (2018). https://doi.org/10.1103/PhysRevLett.121.158101
Young, A.L., et al.: Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with subtype and stage inference. Nat. Commun. 9(1), 4273 (2018)
Zhou, J., Gennatas, E.D., Kramer, J.H., Miller, B.L., Seeley, W.W.: Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 73(6), 1216–1227 (2012). https://doi.org/10.1016/j.neuron.2012.03.004
Acknowledgment
TH, AS and AA are supported by the EPSRC funded UCL Centre for Doctoral Training in Intelligent, Integrated Imaging in Healthcare[EP/S021930/1]; TH, ET and DCA are supported by the Wellcome Trust(221915); DCA and FB are supported by the NIHR Biomedical Research Centre at UCLH and UCL; NPO acknowledges funding from a UKRI Future Leaders Fellowship(MR/S03546X/1).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
He, T. et al. (2023). A Coupled-Mechanisms Modelling Framework for Neurodegeneration. In: Greenspan, H., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14227. Springer, Cham. https://doi.org/10.1007/978-3-031-43993-3_45
Download citation
DOI: https://doi.org/10.1007/978-3-031-43993-3_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43992-6
Online ISBN: 978-3-031-43993-3
eBook Packages: Computer ScienceComputer Science (R0)