Abstract
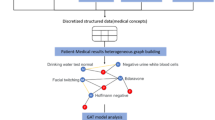
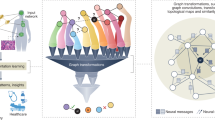
Clinicians prescribe antibiotics by looking at the patient’s health record with an experienced eye. However, the therapy might be rendered futile if the patient has drug resistance. Determining drug resistance requires time-consuming laboratory-level testing while applying clinicians’ heuristics in an automated way is difficult due to the categorical or binary medical events that constitute health records. In this paper, we propose a novel framework for rapid clinical intervention by viewing health records as graphs whose nodes are mapped from medical events and edges as correspondence between events in given a time window. A novel graph-based model is then proposed to extract informative features and yield automated drug resistance analysis from those high-dimensional and scarce graphs. The proposed method integrates multi-task learning into a common feature extracting graph encoder for simultaneous analyses of multiple drugs as well as stabilizing learning. On a massive dataset comprising over 110,000 patients with urinary tract infections, we verify the proposed method is capable of attaining superior performance on the drug resistance prediction problem. Furthermore, automated drug recommendations resemblant to laboratory-level testing can also be made based on the model resistance analysis.
H. Shu and P. Gao—Joint first authors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Attenberg, J., Weinberger, K., Dasgupta, A., Smola, A., Zinkevich, M.: Collaborative email-spam filtering with the hashing trick. In: CEAS (2009)
Bertsimas, D., Van Parys, B.: Sparse high-dimensional regression: exact scalable algorithms and phase transitions. Ann. Stat. 48(1), 300–323 (2020)
Brody, S., Alon, U., Yahav, E.: How attentive are graph attention networks? CoRR abs/2105.14491 (2021)
Chen, Z., Badrinarayanan, V., Lee, C., Rabinovich, A.: GradNorm: gradient normalization for adaptive loss balancing in deep multitask networks. In: Proceedings of the 35th International Conference on Machine Learning, ICML (2018)
Diehl, F.: Edge contraction pooling for graph neural networks. CoRR abs/1905.10990 (2019)
Ferrer, R., et al.: Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program. Crit. Care Med. 42(8), 1749–1755 (2014)
Gao, Y., Ma, J., Zhao, M., Liu, W., Yuille, A.L.: NDDR-CNN: layerwise feature fusing in multi-task CNNs by neural discriminative dimensionality reduction. In: CVPR, pp. 3205–3214 (2019)
Gilmer, J., Schoenholz, S.S., Riley, P.F., Vinyals, O., Dahl, G.E.: Neural message passing for quantum chemistry. In: ICML, pp. 1263–1272 (2017)
Guo, M., Haque, A., Huang, D.-A., Yeung, S., Fei-Fei, L.: Dynamic task prioritization for multitask learning. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11220, pp. 282–299. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01270-0_17
Guo, P., Lee, C., Ulbricht, D.: Learning to branch for multi-task learning. In: Proceedings of the 37th International Conference on Machine Learning, ICML 2020, 13–18 July 2020, Virtual Event. Proceedings of Machine Learning Research, vol. 119, pp. 3854–3863. PMLR (2020)
Hamilton, W.L., Ying, Z., Leskovec, J.: Inductive representation learning on large graphs. In: NeurIPS, pp. 1024–1034 (2017)
Ikram, R., Psutka, R., Carter, A., Priest, P.: An outbreak of multi-drug resistant escherichia coli urinary tract infection in an elderly population: a case-control study of risk factors. BMC Infect. Dis. 15(1), 1–7 (2015)
Kanjilal, S., Oberst, M., Boominathan, S., Zhou, H., Hooper, D.C., Sontag, D.A.: A decision algorithm to promote outpatient antimicrobial stewardship for uncomplicated urinary tract infection. Science Transl. Med. 12 (2020)
Kendall, A., Gal, Y., Cipolla, R.: Multi-task learning using uncertainty to weigh losses for scene geometry and semantics. In: 2018 IEEE Conference on Computer Vision and Pattern Recognition, CVPR (2018)
Kipf, T.N., Welling, M.: Semi-supervised classification with graph convolutional networks. In: ICLR (2017)
Kokkinos, I.: UberNet: training a universal convolutional neural network for low-, mid-, and high-level vision using diverse datasets and limited memory. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition, CVPR (2017)
Lee, J., Lee, I., Kang, J.: Self-attention graph pooling. In: Proceedings of the 36th International Conference on Machine Learning, ICML 2019, Long Beach, California, USA, 9–15 June 2019. Proceedings of Machine Learning Research, vol. 97, pp. 3734–3743. PMLR (2019)
Lin, T., Goyal, P., Girshick, R.B., He, K., Dollár, P.: Focal loss for dense object detection. In: ICCV, pp. 2999–3007 (2017)
Liu, S., Johns, E., Davison, A.J.: End-to-end multi-task learning with attention. In: CVPR, pp. 1871–1880 (2019)
Ma, Y., Wang, S., Aggarwal, C.C., Tang, J.: Graph convolutional networks with eigenpooling. In: ACM SIGKDD, pp. 723–731 (2019)
Michael, O., Soorajnath, B., Helen, Z., Sanjat, K., Sontag, D.: AMR-UTI: antimicrobial resistance in urinary tract infections (version 1.0.0). PhysioNet (2020)
Misra, I., Shrivastava, A., Gupta, A., Hebert, M.: Cross-stitch networks for multi-task learning. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2016, Las Vegas, NV, USA, 27–30 June 2016, pp. 3994–4003. IEEE Computer Society (2016)
Reller, L.B., Weinstein, M., Jorgensen, J.H., Ferraro, M.J.: Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49(11), 1749–1755 (2009)
Seger, C.: An investigation of categorical variable encoding techniques in machine learning: binary versus one-hot and feature hashing (2018)
Sener, O., Koltun, V.: Multi-task learning as multi-objective optimization. In: NeurIPS, pp. 525–536 (2018)
Shervashidze, N., Schweitzer, P., van Leeuwen, E.J., Mehlhorn, K., Borgwardt, K.M.: Weisfeiler-Lehman graph kernels. J. Mach. Learn. Res. 12, 2539–2561 (2011)
Tulyakov, S., Farooq, F., Mansukhani, P., Govindaraju, V.: Symmetric hash functions for secure fingerprint biometric systems. Pattern Recogn. Lett. 28(16), 2427–2436 (2007)
Vandenhende, S., Georgoulis, S., Gansbeke, W.V., Proesmans, M., Dai, D., Gool, L.V.: Multi-task learning for dense prediction tasks: a survey. IEEE Trans. Pattern Anal. Mach. Intell. 44(7), 3614–3633 (2022)
Velickovic, P., Cucurull, G., Casanova, A., Romero, A., Liò, P., Bengio, Y.: Graph attention networks. In: ICLR (2018)
Ventola, C.L.: The antibiotic resistance crisis: part 1: causes and threats. Pharm. Ther. 40(4), 277 (2015)
Wu, F., Souza, A., Zhang, T., Fifty, C., Yu, T., Weinberger, K.Q.: Simplifying graph convolutional networks. In: ICML, pp. 6861–6871 (2019)
Wu, Z., Pan, S., Chen, F., Long, G., Zhang, C., Yu, P.S.: A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 32, 4–24 (2019)
Xu, K., Hu, W., Leskovec, J., Jegelka, S.: How powerful are graph neural networks? In: ICLR (2019)
Yelin, I., et al.: Personal clinical history predicts antibiotic resistance of urinary tract infections. Nat. Med. 25(7), 1143–1152 (2019)
Ying, Z., You, J., Morris, C., Ren, X., Hamilton, W.L., Leskovec, J.: Hierarchical graph representation learning with differentiable pooling. In: NeurIPS, pp. 4805–4815 (2018)
Zhang, M.L., Zhou, Z.H.: A review on multi-label learning algorithms. IEEE Trans. Knowl. Data Eng. 26, 1819–1837 (2014)
Acknowledgement
This work was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research Number JP21H03446, NICT 03501, JST-AIP JPMJCR21U4.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Shu, H., Gao, P., Zhu, L., Chen, Z., Matsubara, Y., Sakurai, Y. (2023). Drugs Resistance Analysis from Scarce Health Records via Multi-task Graph Representation. In: Yang, X., et al. Advanced Data Mining and Applications. ADMA 2023. Lecture Notes in Computer Science(), vol 14178. Springer, Cham. https://doi.org/10.1007/978-3-031-46671-7_8
Download citation
DOI: https://doi.org/10.1007/978-3-031-46671-7_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-46670-0
Online ISBN: 978-3-031-46671-7
eBook Packages: Computer ScienceComputer Science (R0)