Abstract
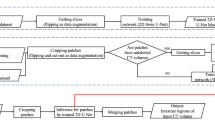
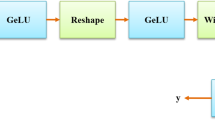
The intestine is an essential digestive organ that can cause serious health problems once diseased. This paper proposes a method for intestine segmentation to intestine obstruction diagnosis assistance called multi-dimensional U-Net (M U-Net). We employ two encoders to extract features from two-dimensional (2D) CT slices and three-dimensional (3D) CT patches. These two encoders collaborate to enhance the segmentation accuracy of the model. Additionally, we incorporate deep supervision with the M U-Net to reduce the limitation of training with sparse label data sets. The experimental results demonstrated that the Dice of the proposed method was 73.22%, the recall was 79.89%, and the precision was 70.61%.
Q. An—Contributing author.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sinicrope, F., Ileus and Bowel Obstruction: Holland-Frei Cancer Medicine, 6th edn. Hamilton BC Decker, Hamilton (2003)
Roth, H.R., et al.: Deep learning and its application to medical image segmentation. Med. Imag. Technol. 36(2), 63–71 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015, Part III. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Xiao, X., Lian, S., Luo, Z., Li, S.: Weighted Res-UNet for high-quality retina vessel segmentation. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327–331. IEEE (2018)
Cai, S., Tian, Y., Lui, H., Zeng, H., Wu, Y., Chen, G.: Dense-UNet: a novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant. Imag. Med. Surg. 10(6) (2020). https://qims.amegroups.com/article/view/43519. ISSN 2223–4306
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016, Part II. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Angermann, C., Haltmeier, M.: Random 2.5 D U-Net for Fully 3D Segmentation, pp. 158–166 (2019)
Han, X.: Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv preprint arXiv:1704.07239 (2017)
Lv, P., Wang, J., Wang, H.: 2.5D lightweight RIU-Net for automatic liver and tumor segmentation from CT. Biomed. Signal Process. Control 75, 103567 (2022). https://doi.org/10.1016/j.bspc.2022.103567. https://www.sciencedirect.com/science/article/pii/S1746809422000891. ISSN 1746–8094
Li, X., Chen, H., Qi, X., Dou, Q., Chi-Wing, F., Heng, P.-A.: H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 37(12), 2663–2674 (2018)
Rajamani, K., et al.: Segmentation of colon and removal of opacified fluid for virtual colonoscopy. Pattern Anal. Appl. 21(1), 205–219 (2018)
Zhang, W., Kim, H.M.: Fully automatic colon segmentation in computed tomography colonography. In: 2016 IEEE International Conference on Signal and Image Processing (ICSIP), pp. 51–55. IEEE (2016)
Sato, Y., et al.: Tissue classification based on 3D local intensity structures for volume rendering. IEEE Trans. Visual Comput. Graphics 6(2), 160–180 (2000)
Frimmel, H., Näppi, J., Yoshida, H.: Centerline-based colon segmentation for CT colonography. Med. Phys. 32(8), 2665–2672 (2005)
Procedures, I.-G., Barr, K., Laframboise, J., Ungi, T., Hookey, L., Fichtinger, G.: Automated segmentation of computed tomography colonography images using a 3D U-Net. In: SPIE Medical Imaging 2020. Robotic Interventions, and Modeling, vol. 1315, pp. 635–641 (2020)
Shin, S.Y., Lee, S., Elton, D., Gulley, J.L., Summers, R.M.: Deep small bowel segmentation with cylindrical topological constraints. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12264, pp. 207–215. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59719-1_21
Oda, H., et al.: Visualizing intestines for diagnostic assistance of ileus based on intestinal region segmentation from 3D CT images. In: SPIE Medical Imaging 2020: Computer-Aided Diagnosis, vol. 11314, pp. 728–735 (2020)
Zeng, Y., Tsui, P.-H., Weiwei, W., Zhou, Z., Shuicai, W.: Fetal ultrasound image segmentation for automatic head circumference biometry using deeply supervised attention-gated V-Net. J. Digit. Imaging 34(1), 134–148 (2021)
Zhou, Y., Xie, L., Fishman, E.K., Yuille, A.L.: Deep supervision for pancreatic cyst segmentation in abdominal CT scans. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017, Part III. LNCS, vol. 10435, pp. 222–230. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_26
Milletari, F., Navab, N., Ahmadi., S.-A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Karimi, D., Salcudean, S.E.: Reducing the Hausdorff distance in medical image segmentation with convolutional neural networks. IEEE Trans. Med. Imaging 39(2), 499–513 (2019)
Acknowledgments
Thanks for the help and advice from Mori laboratory. A part of this research was supported by Hori Sciences and Arts Foundation, MEXT/JSPS KAKENHI (17H00867, 22H03203), the JSPS Bilateral International Collaboration Grants, and the JST CREST (JPMJCR20D5). And this work was also financially supported by the JST SPRING, Grant Number JPMJSP2125.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
An, Q. et al. (2024). M U-Net: Intestine Segmentation Using Multi-dimensional Features for Ileus Diagnosis Assistance. In: Wu, S., Shabestari, B., Xing, L. (eds) Applications of Medical Artificial Intelligence. AMAI 2023. Lecture Notes in Computer Science, vol 14313. Springer, Cham. https://doi.org/10.1007/978-3-031-47076-9_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-47076-9_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-47075-2
Online ISBN: 978-3-031-47076-9
eBook Packages: Computer ScienceComputer Science (R0)