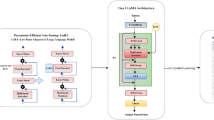
Abstract
Foundation large language models (LLMs) have shown an impressive ability to solve tasks across a wide range of fields including health. To effectively solve personalized health tasks, LLMs need the ability to ingest a diversity of data modalities that are relevant to an individual’s health status. In this paper, we take a step towards creating multimodal LLMs for health that are grounded in individual-specific data by developing a framework (HeLM: Health Large Language Model for Multimodal Understanding) that enables LLMs to use high-dimensional clinical modalities to estimate underlying disease risk. HeLM encodes complex data modalities by learning an encoder that maps them into the LLM’s token embedding space and for simple modalities like tabular data by serializing the data into text. Using data from the UK Biobank, we show that HeLM can effectively use demographic and clinical features in addition to high-dimensional time-series data to estimate disease risk. For example, HeLM achieves an AUROC of 0.75 for asthma prediction when combining tabular and spirogram data modalities compared with 0.49 when only using tabular data. Overall, we find that HeLM outperforms or performs at parity with classical machine learning approaches across a selection of eight binary traits. Furthermore, we investigate the downstream uses of this model such as its generalizability to out-of-distribution traits and its ability to power conversations around individual health and wellness.
A. Belyaeva and J. Cosentino—Equal contribution.
C.Y. McLean and N.A. Furlotte—Equal supervision.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Acosta, J.N., Falcone, G.J., Rajpurkar, P., Topol, E.J.: Multimodal biomedical AI. Nat. Med. 28(9), 1773–1784 (2022)
Alayrac, J.B., et al.: Flamingo: a visual language model for few-shot learning. In: Advances in Neural Information Processing Systems, vol. 35, pp. 23716–23736 (2022)
Alipanahi, B., et al.: Large-scale machine-learning-based phenotyping significantly improves genomic discovery for optic nerve head morphology. Am. J. Hum. Genet. 108(7), 1217–1230 (2021)
Brown, T., et al.: Language models are few-shot learners. In: Advances in Neural Information Processing Systems, vol. 33, pp. 1877–1901 (2020)
Bycroft, C., et al.: The UK Biobank resource with deep phenotyping and genomic data. Nature 562(7726), 203–209 (2018)
Chen, T., Guestrin, C.: XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD 2016, pp. 785–794. ACM, New York (2016). https://doi.org/10.1145/2939672.2939785
Chung, H.W., et al.: Scaling instruction-finetuned language models. arXiv preprint arXiv:2210.11416 (2022)
Cosentino, J., et al.: Inference of chronic obstructive pulmonary disease with deep learning on raw spirograms identifies new genetic loci and improves risk models. Nat. Genet. 55, 787–795 (2023)
Diaz-Papkovich, A., Anderson-Trocmé, L., Ben-Eghan, C., Gravel, S.: UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS Genet. 15(11), e1008432 (2019)
Dinh, T., et al.: LIFT: language-interfaced fine-tuning for non-language machine learning tasks. In: Advances in Neural Information Processing Systems, vol. 35, pp. 11763–11784 (2022)
Driess, D., et al.: PaLM-E: an embodied multimodal language model. arXiv preprint arXiv:2303.03378 (2023)
Girdhar, R., et al.: ImageBind: one embedding space to bind them all. arXiv preprint arXiv:2305.05665 (2023)
Google: PaLM 2 technical report. arXiv preprint arXiv:2305.10403 (2023)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
He, T., Zhang, Z., Zhang, H., Zhang, Z., Xie, J., Li, M.: Bag of tricks for image classification with convolutional neural networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 558–567 (2019)
Hegselmann, S., Buendia, A., Lang, H., Agrawal, M., Jiang, X., Sontag, D.: TabLLM: few-shot classification of tabular data with large language models. In: International Conference on Artificial Intelligence and Statistics, pp. 5549–5581. PMLR (2023)
Jia, C., et al.: Scaling up visual and vision-language representation learning with noisy text supervision. In: International Conference on Machine Learning, pp. 4904–4916. PMLR (2021)
Kirk, H.R., Vidgen, B., Röttger, P., Hale, S.A.: Personalisation within bounds: a risk taxonomy and policy framework for the alignment of large language models with personalised feedback. arXiv preprint arXiv:2303.05453 (2023)
Kline, A., et al.: Multimodal machine learning in precision health: a scoping review. npj Digit. Med. 5(1), 171 (2022)
Lester, B., Al-Rfou, R., Constant, N.: The power of scale for parameter-efficient prompt tuning. In: Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, pp. 3045–3059 (2021). https://doi.org/10.18653/v1/2021.emnlp-main.243
Li, J., Li, D., Savarese, S., Hoi, S.: BLIP-2: bootstrapping language-image pre-training with frozen image encoders and large language models. arXiv preprint arXiv:2301.12597 (2023)
Lu, J., Clark, C., Zellers, R., Mottaghi, R., Kembhavi, A.: Unified-IO: a unified model for vision, language, and multi-modal tasks. arXiv preprint arXiv:2206.08916 (2022)
Moor, M., et al.: Foundation models for generalist medical artificial intelligence. Nature 616(7956), 259–265 (2023)
OpenAI: GPT-4 technical report. arXiv preprint arXiv:2303.08774 (2023)
Radford, A., et al.: Learning transferable visual models from natural language supervision. In: International Conference on Machine Learning, pp. 8748–8763. PMLR (2021)
Recht, B., Roelofs, R., Schmidt, L., Shankar, V.: Do CIFAR-10 classifiers generalize to CIFAR-10? arXiv preprint arXiv:1806.00451 (2018)
Sakornsakolpat, P., et al.: Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat. Genet. 51(3), 494–505 (2019)
Salemi, A., Mysore, S., Bendersky, M., Zamani, H.: LaMP: when large language models meet personalization. arXiv preprint arXiv:2304.11406 (2023)
Shrine, N., et al.: New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 51(3), 481–493 (2019)
Singhal, K., et al.: Large language models encode clinical knowledge. arXiv preprint arXiv:2212.13138 (2022)
Singhal, K., et al.: Towards expert-level medical question answering with large language models. arXiv preprint arXiv:2212.13138 (2022)
Steinberg, E., Jung, K., Fries, J.A., Corbin, C.K., Pfohl, S.R., Shah, N.H.: Language models are an effective representation learning technique for electronic health record data. J. Biomed. Inform. 113, 103637 (2021)
Vestbo, J., et al.: Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 187(4), 347–365 (2013)
Vokinger, K.N., Feuerriegel, S., Kesselheim, A.S.: Mitigating bias in machine learning for medicine. Commun. Med. 1(1), 25 (2021)
Wang, Y., et al.: Preserving in-context learning ability in large language model fine-tuning. arXiv preprint arXiv:2211.00635 (2022)
Wei, J., et al.: Emergent abilities of large language models. arXiv preprint arXiv:2206.07682 (2022)
Yang, K.D., et al.: Multi-domain translation between single-cell imaging and sequencing data using autoencoders. Nat. Commun. 12(1), 31 (2021)
Yang, X., et al.: A large language model for electronic health records. npj Digit. Med. 5(1), 194 (2022)
Yu, J., Wang, Z., Vasudevan, V., Yeung, L., Seyedhosseini, M., Wu, Y.: CoCa: contrastive captioners are image-text foundation models. arXiv preprint arXiv:2205.01917 (2022)
Zhou, H.Y., Chen, X., Zhang, Y., Luo, R., Wang, L., Yu, Y.: Generalized radiograph representation learning via cross-supervision between images and free-text radiology reports. Nat. Mach. Intell. 4(1), 32–40 (2022)
Acknowledgements
The authors would like to thank Katrin Tomanek for providing software, inspiration, and know-how that influenced the direction of this work. We also thank Ted Yun for helpful discussions and feedback.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Belyaeva, A. et al. (2024). Multimodal LLMs for Health Grounded in Individual-Specific Data. In: Maier, A.K., Schnabel, J.A., Tiwari, P., Stegle, O. (eds) Machine Learning for Multimodal Healthcare Data. ML4MHD 2023. Lecture Notes in Computer Science, vol 14315. Springer, Cham. https://doi.org/10.1007/978-3-031-47679-2_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-47679-2_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-47678-5
Online ISBN: 978-3-031-47679-2
eBook Packages: Computer ScienceComputer Science (R0)