Abstract
The analysis of fundus images for the early screening of eye diseases is of great clinical importance. Traditional methods for such analysis are time-consuming and expensive as it requires a trained clinician. Therefore, the need for a comprehensive and automated method of retinal image screening to diagnose and grade retinal diseases has long been recognized. In the past decade, with the substantial developments in computer vision and deep learning, machine learning methods became highly effective in this field as clinical-decision support methods. However, most of these algorithms face challenges like computational feasibility, reliability, and interpretability.
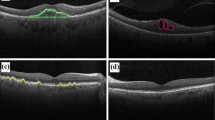
In this paper, we develop a novel approach to this crucial task in retinal image screening. By employing topological data analysis tools, for the most common retinal diseases, i.e., Diabetic Retinopathy (DR), Glaucoma, and Age-related Macular Degeneration (AMD), we observe different types of topological patterns between normal and abnormal classes. These patterns enable us to produce topological fingerprints of fundus images, and we use them as feature vectors with standard ML methods. Our computationally efficient model ToFi-ML outperforms or gives highly competitive accuracy results with state-of-the-art deep learning methods. Furthermore, our topological fingerprints are both explainable and interpretable, and can easily be integrated with future early-screening ML models.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Adcock, A., Rubin, D., Carlsson, G.: Classification of hepatic lesions using the matching metric. Comput. Vis. Image Underst. 121, 36–42 (2014)
Ajitha, S., Akkara, J.D., Judy, M.: Identification of glaucoma from fundus images using deep learning techniques. Indian J. Ophthalmol. 69(10), 2702 (2021)
APTOS: Asia Pacific Tele-Ophthalmology Society (APTOS) 2019 Blindness Detection Dataset (2019). https://www.kaggle.com/c/aptos2019-blindness-detection
Bendich, P., Marron, J.S., Miller, E., Pieloch, A., Skwerer, S.: Persistent homology analysis of brain artery trees. Ann. Appl. Stat. 10(1), 198 (2016)
Berry, E., Chen, Y.C., Cisewski-Kehe, J., Fasy, B.T.: Functional summaries of persistence diagrams. J. Appl. Comput. Topol. 4(2), 211–262 (2020)
Bodapati, J.D., et al.: Blended multi-modal deep convnet features for diabetic retinopathy severity prediction. Electronics 9(6), 914 (2020)
Bodapati, J.D., et al.: Composite deep neural network with gated-attention mechanism for DR severity classification. J. Amb. Int. Hum. Compt. 12(10), 9825–9839 (2021)
Cámara, P.G., Levine, A.J., Rabadan, R.: Inference of ancestral recombination graphs through topological data analysis. PLoS Comput. Biol. 12(8), e1005071 (2016)
Campbell, M.J., Machin, D., Walters, S.J.: Medical Statistics: A Textbook for the Health Sciences. Wiley, Hoboken (2010)
Carlsson, G., Vejdemo-Johansson, M.: Topological Data Analysis with Applications. Cambridge University Press, Cambridge (2021)
Chakraborty, R., Pramanik, A.: DCNN-based prediction model for detection of AMD from color fundus images. Med. Bio. Eng. Comput. 60(5), 1431–1448 (2022)
Chaturvedi, S.S., Gupta, K., Ninawe, V., Prasad, P.S.: Automated diabetic retinopathy grading using deep convolutional neural network. arXiv preprint arXiv:2004.06334 (2020)
Chazal, F., Michel, B.: An introduction to topological data analysis: fundamental and practical aspects for data scientists. Front. Artif. Intell. 4, 108 (2021)
Chen, T., Kornblith, S., Norouzi, M., Hinton, G.: A simple framework for contrastive learning of visual representations. In: ICML, pp. 1597–1607. PMLR (2020)
Crawford, L., et al.: Predicting clinical outcomes in glioblastoma: an application of topological and functional data analysis. J. Am. Stat. Assoc. 115(531), 1139–1150 (2020)
Dey, T.K., Wang, Y.: Computational Topology for Data Analysis. Cambridge University Press, Cambridge (2022)
Elangovan, P., Nath, M.K.: Glaucoma assessment from color fundus images using convolutional neural network. Int. J. Imaging Syst. Technol. 31(2), 955–971 (2021)
Fang, H., et al.: ADAM challenge: detecting age-related macular degeneration from fundus images. IEEE Trans. Med. Imaging 41, 2828–2847 (2022)
Fawcett, T.: An introduction to ROC analysis. Pattern Rec. Lett. 27(8), 861–874 (2006)
Feng, Z., Xu, C., Tao, D.: Self-supervised representation learning by rotation feature decoupling. In: CVPR, pp. 10364–10374 (2019)
Fernández, A., et al.: SMOTE for learning from imbalanced data. J. Artif. Intell. Res. 61, 863–905 (2018)
Fu, H., et al.: ADAM: automatic detection challenge on AMD (2020). https://doi.org/10.21227/dt4f-rt59
Giansiracusa, N., Giansiracusa, R., Moon, C.: Persistent homology machine learning for fingerprint classification. In: ICMLA, pp. 1219–1226. IEEE (2019)
Gibbons, J.D., Chakraborti, S.: Nonparametric Statistical Inference. CRC Press, Boca Raton (2014)
Giunti, B.: TDA applications library (2022). https://www.zotero.org/groups/2425412/tda-applications/library
Goutam, B., Hashmi, M.F., Geem, Z.W., Bokde, N.D.: A comprehensive review of deep learning strategies in retinal disease diagnosis using fundus images. IEEE Access 10, 57796–57823 (2022)
Greenspan, H., Van Ginneken, B., Summers, R.M.: Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 35(5), 1153–1159 (2016)
GUDHI: The GUDHI project (2020). https://gudhi.inria.fr/doc/3.3.0/
Hatcher, A.: Algebraic Topology. Cambridge University Press, Cambridge (2002)
Islam, M.R., et al.: Applying supervised contrastive learning for the detection of DR and its severity levels from fundus images. Comput. Biol. Med. 146, 105602 (2022)
Joblove, G.H., Greenberg, D.: Color spaces for computer graphics. In: Proceedings of the 5th Annual Conference on Computer Graphics and Interactive Techniques, pp. 20–25 (1978)
Kanari, L., et al.: A topological representation of branching neuronal morphologies. Neuroinformatics 16(1), 3–13 (2018)
Kumar, G., Chatterjee, S., Chattopadhyay, C.: DRISTI: a hybrid deep neural network for diabetic retinopathy diagnosis. Signal Image Video Process. 15(8), 1679–1686 (2021)
Latif, J., Tu, S., Xiao, C., Ur Rehman, S., Imran, A., Latif, Y.: ODGNet: a deep learning model for automated optic disc localization and glaucoma classification using fundus images. SN Appl. Sci. 4(4), 1–11 (2022)
Li, T., et al.: Applications of deep learning in fundus images: a review. Med. Image Anal. 69, 101971 (2021)
Li, X., et al.: Rotation-oriented collaborative self-supervised learning for retinal disease diagnosis. IEEE Trans. Med. Imaging 40(9), 2284–2294 (2021)
Liao, W., et al.: Clinical interpretable deep learning model for glaucoma diagnosis. IEEE J. Biomed. Health Inform. 24(5), 1405–1412 (2019)
Macsik, P., Pavlovicova, J., Goga, J., Kajan, S.: Local binary CNN for diabetic retinopathy classification on fundus images. Acta Polytech. Hung. 19(7), 27–45 (2022)
Milosavljević, N., Morozov, D., Skraba, P.: Zigzag persistent homology in matrix multiplication time. In: SoCG, pp. 216–225 (2011)
Orlando, J.I., et al.: An ensemble deep learning based approach for red lesion detection in fundus images. Comput. Methods Programs Biomed. 153, 115–127 (2018)
Otter, N., Porter, M.A., Tillmann, U., Grindrod, P., Harrington, H.A.: A roadmap for the computation of persistent homology. EPJ Data Sci. 6, 1–38 (2017)
Pedregosa, F., et al.: Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Pratt, H., Coenen, F., Broadbent, D.M., Harding, S.P., Zheng, Y.: Convolutional neural networks for diabetic retinopathy. Procedia Comput. Sci. 90, 200–205 (2016)
Rieck, B., et al.: Uncovering the topology of time-varying fmri data using cubical persistence. In: NeurIPS, vol. 33, pp. 6900–6912 (2020)
Sarhan, M.H., et al.: Machine learning techniques for ophthalmic data processing: a review. IEEE J. Biomed. Health Inform. 24(12), 3338–3350 (2020)
Skaf, Y., Laubenbacher, R.: Topological data analysis in biomedicine: a review. J. Biomed. Inform. 104082 (2022)
Skraba, P., Ovsjanikov, M., Chazal, F., Guibas, L.: Persistence-based segmentation of deformable shapes. In: CVPR-Workshops, pp. 45–52. IEEE (2010)
Srivastava, O., et al.: Artificial intelligence and machine learning in ophthalmology: a review. Indian J. Ophthalmol. 71(1), 11–17 (2023)
Stolz, B.J., et al.: Topological data analysis of task-based fMRI data from experiments on schizophrenia. J. Phys.: Complexity 2(3), 035006 (2021)
Taj, I.A., et al.: An ensemble framework based on deep CNNs for glaucoma classification. Math. Biosci. Eng. 18(5), 5321–5347 (2021)
Tauzin, G., et al.: giotto-TDA: a TDA toolkit for machine learning and data exploration (2020)
Ting, D.S.W., et al.: Artificial intelligence and deep learning in ophthalmology (2019)
Ting, D.S., et al.: Deep learning in ophthalmology: the technical and clinical considerations. Prog. Retin. Eye Res. 72, 100759 (2019)
Wasserman, L.: Topological data analysis. Annu. Rev. Stat. Appl. 5, 501–532 (2018)
World Health Organization, W.: World vision report (2019). https://www.who.int/publications/i/item/9789241516570
Wu, Z., Xiong, Y., Yu, S.X., Lin, D.: Unsupervised feature learning via non-parametric instance discrimination. In: CVPR, pp. 3733–3742 (2018)
Ye, M., Zhang, X., Yuen, P.C., Chang, S.F.: Unsupervised embedding learning via invariant and spreading instance feature. In: CVPR, pp. 6210–6219 (2019)
Zhang, Z., et al.: ORIGA-light: an online retinal fundus image database for glaucoma analysis and research. In: 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, pp. 3065–3068. IEEE (2010)
Zhang, Z., Ji, Z., Chen, Q., Yuan, S., Fan, W.: Joint optimization of CycleGAN and CNN classifier for detection and localization of retinal pathologies on color fundus photographs. IEEE J. Biomed. Health Inform. 26(1), 115–126 (2021)
Zhao, X., et al.: Glaucoma screening pipeline based on clinical measurements and hidden features. IET Image Process. 13(12), 2213–2223 (2019)
Acknowledgements
This work was partially supported by National Science Foundation (Grant # DMS-2202584) and by Simons Foundation (Grant # 579977).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Ahmed, F., Coskunuzer, B. (2024). ToFi-ML: Retinal Image Screening with Topological Machine Learning. In: Waiter, G., Lambrou, T., Leontidis, G., Oren, N., Morris, T., Gordon, S. (eds) Medical Image Understanding and Analysis. MIUA 2023. Lecture Notes in Computer Science, vol 14122. Springer, Cham. https://doi.org/10.1007/978-3-031-48593-0_21
Download citation
DOI: https://doi.org/10.1007/978-3-031-48593-0_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-48592-3
Online ISBN: 978-3-031-48593-0
eBook Packages: Computer ScienceComputer Science (R0)